The golgi apparatus
Servier Medical Art. Licensed under CC BY 2.0.
Sometimes science takes a village
Alexandra Navarro, a graduate student in Whitehead Institute Member Iain Cheeseman’s lab, was studying the gene for CENPR, a protein related to cell division—the Cheeseman lab’s research focus—when she came across something interesting: another molecule hidden in CENPR’s genetic code. The hidden molecule is a peptide only 37 amino acids long, too small to show up in most surveys of the cell. It gets created only when the genetic code for CENPR is translated from an offset start and stopping place—essentially, when a cell reads the instructions for making CENPR in a different way. The Cheeseman lab has become very interested in these sorts of hidden molecules, which they have found lurking in a number of other molecules’ genetic codes. Navarro began studying the peptide as a side project during slow periods in her main research on cell division proteins. However, as her research on the peptide progressed, Navarro eventually found herself unsure of how to proceed. CENPR belongs in the centromere, a part of the cell necessary for cell division, but the alternative peptide ends up in the Golgi, a structure that helps to modify molecules and prep them for delivery to different destinations. In other words, the peptide had nothing to do with the part of the cell that Navarro and Cheeseman typically study.
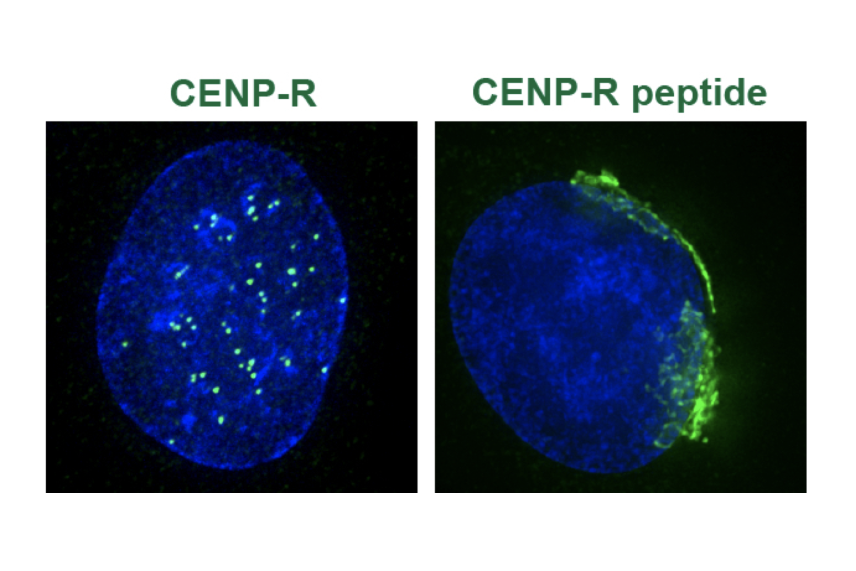
The peptide hidden in CENPR's genetic code localizes to a completely different part of the cell, as seen here. CENPR is on the left, and the peptide on the right, both in green.
Alexandra Navarro
Usually when Navarro comes across something outside of her area of expertise, she will consult with her lab mates, others in Whitehead Institute, or nearby collaborators. However, none of her usual collaborators' research focuses on the Golgi, so this time Cheeseman suggested that Navarro share what they had found and ask for input from as wide a circle of researchers as possible—on the internet. Often, researchers guard their work in progress carefully, reluctant to share it lest they be scooped, which means someone else publishes a paper on the same topic first. In the competitive world of academic research, where publishing papers is a key part of getting jobs, tenure, and future funding, the specter of scooping can loom large. But science is also an inherently collaborative practice, with scientists contributing droplets of discovery to a shared pool of knowledge, so that new findings can be built upon what came before. Cheeseman is a board member of ASAPbio (Accelerating Science and Publication in biology), a nonprofit that promotes open communication, the use of preprints, and transparent peer review in the life sciences. Researchers like Cheeseman believe that if science adopts more transparent and collaborative practices, such as more frequently and widely sharing research in progress, this will benefit both the people involved and the quality of the science, and will speed up the search for discoveries with the potential to positively impact humankind. But how helpful are such “open science” practices in reality? Navarro and Cheeseman had the perfect opportunity to find out.
The power of preprints
Navarro and Cheeseman wrote up what they knew so far–they had found a hidden peptide that localizes exclusively to the Golgi, and it stays there throughout the cell cycle–as a “preprint in progress,” an incomplete draft of a paper that acknowledges there is more to come. In December 2020, they posted the preprint in progress to bioRxiv, a website that serves as a repository for biology preprints, or papers that have not yet

been published. The site was inspired by arXiv, a similar repository launched in 1991 to provide free and easy access to research in math, physics, computer science, and similar fields. arXiv has become a central hub for research in these fields, with an average of 10-15,000 submissions and 30 million downloads per month. The biology fields were slower to create such a hub: BioRxiv launched in 2013. In December, 2021, it received around 3,000 submissions and 2.3 million downloads.
Navarro and Cheeseman’s decision to post a preprint in progress to bioRvix is not common practice, but a lot of researchers have started posting preprints that resemble the final paper closer to publication. Some journals even require it. This type of early sharing has many benefits: contrary to the fear that sharing research before publication will lead to scooping, it allows researchers to stake a claim sooner by making their work public record pre-publication. Preprints enable researchers to show off their most current work during the narrow windows of the academic job cycle. This can be particularly crucial for early career researchers whose biggest project to date—such as graduate thesis work—is still in publication limbo. Preprints also allow new ideas and knowledge to get out into the world sooner, the better to inspire other researchers. Findings that seemed minor at first have provided the key insight for someone else’s major discovery throughout biology’s history. The sooner research is shared, the sooner it can be built upon to develop important advances, like new medicines or a better model of how a disease spreads.
Navarro and Cheeseman weren’t expecting their discovery to have that kind of major impact, but they knew the peptide could be useful to researchers studying the Golgi. The peptide is small and doesn’t disrupt any functions in the cell. Researchers can attach fluorescent proteins to it that make the Golgi glow in imaging. These traits make the peptide a useful potential tool. Since Navarro and Cheeseman posted the preprint, multiple researchers have reached out about using the peptide.

However, the main goal of posting a preprint in progress, as opposed to a polished preprint, is to ask for input to further the research. The morning after the researchers put their preprint on bioRxiv, Cheeseman shared it on Twitter and asked for feedback. Other researchers soon shared the tweet further, and responses started flooding in. Some researchers simply commented that they found the project interesting, which was reassuring for Cheeseman and Navarro.
“It was nice to see that we weren't the only ones who thought this thing we found was really cool,” Navarro says. “It gave me a lot of motivation to keep moving on with this project.”
Then, some researchers had specific questions and ideas. The topic that seemed of greatest interest was how the peptide ends up at the Golgi, followed by where exactly in the Golgi it ends up. Researchers suggested online tools that might help predict answers to these questions. They proposed different mechanisms that might be involved.
Navarro used these suggestions to design a new series of experiments, in order to better characterize how the peptide associates with the Golgi. She found out that the peptide attaches to the Golgi’s outer-facing membrane. She started developing an understanding of which of peptide’s 37 amino acids were necessary for Golgi localization, and so was able to narrow in on a 14-amino acid sequence within the peptide that was sufficient for this localization.
Her next question was what specific mechanisms were driving the peptide’s Golgi localization. Navarro had a good lead for one mechanism: the evidence and outside input suggested that after the peptide was created, it likely underwent a modification that gave it a sticky tag to anchor it to the Golgi. What would be the best experiments to confirm this mechanism and determine the other mechanisms involved? Navarro and Cheeseman decided it was time to check back in with the crowd online.
Narrowing in on answers
Navarro and Cheeseman updated their preprint with their new findings, and invited further feedback. This time, they had a specific ask: how to test whether the peptide has the modification they suspected. They received suggestions: a probe, an inhibitor. They also received some unexpected feedback that took them in a new direction. Harmit Malik, professor and associate director of the basic sciences division at Fred Hutch, studies the evolutionary changes that occur in genes. Malik found the peptide interesting enough to dig into its evolutionary history across primates. He emailed Cheeseman and Navarro his findings. Versions of the peptide existed in many primates, and some of the variations between species affected where the peptide ended up. This was a rich new vein of inquiry for Navarro to follow in order to pinpoint exactly which parts of the sequence were necessary for Golgi localization, and the researchers might never have come across it if they had not sought input online.
Guided by the latest set of suggestions, Navarro resumed work on the project. She found evidence that the peptide does undergo the suspected modification. She winnowed down to a 10-amino acid sequence within the peptide that appears to be the minimal sequence necessary for this type of Golgi localization. Navarro and Cheeseman rewrote the paper, adding the discovery of a minimal Golgi targeting sequence—basically a postal code that marks a molecule’s destination as the Golgi. They posted a third version of the preprint in September, 2021. This time, Cheeseman did not ask Twitter for feedback: the paper may undergo more changes, but it now contains a complete research story.
The changing face of science
Based on their experience, would Cheeseman and Navarro recommend sharing preprints in progress? The answer is a resounding yes—if the project is a good fit. Both agree that for projects like this, where the subject is outside the expertise of a researcher’s usual circle of collaborators, asking the wider scientific community for help can be extremely valuable.
“I often share my research with other people at Whitehead Institute, and other cell division researchers at conferences, but this process allowed me to share it with people who work in different scientific areas, with whom I would not normally engage,” Navarro says.
Cheeseman hopes that sharing hubs like bioRxiv will develop ways for even larger and more diverse groups of scientists to connect.
If researchers are hesitant to use an open science approach, Cheeseman and Navarro recommend testing the waters by starting with a lower stakes project. In this case, Navarro’s Golgi paper was a side project, something of personal interest but not integral to her career. Having had a positive experience using an open approach on this project, Cheeseman and Navarro agree they would be comfortable using such an approach again in the future.
“I wouldn’t suggest sharing a preprint in progress for every paper, but I think constructive opportunities are more plentiful than researchers may realize,” Cheeseman says.
In general, Cheeseman thinks, the biology field needs to re-envision how its science gets shared.
“The idea that one size fits all, that everything needs to be a multi-figure paper in a high impact journal, is just not compatible with the way that people do research,” Cheeseman says. “We need to get flexible and explore and value scholarship in every form.”
As for the peptide paper? Regardless of where it ends up, Cheeseman and Navarro consider their open science experiment a success. By sharing their research and asking for input, they gained insights, research tools, and points of view that took the project from a curious finding to a rich understanding of the mechanisms behind Golgi localization. Their early realization that the peptide functions outside of their region of expertise could have been a dead end. But by being open about what they were working on and what sort of guidance they needed, the researchers were able to overcome that hurdle and decode their mystery peptide, with a little help from the wider scientific community.
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu


