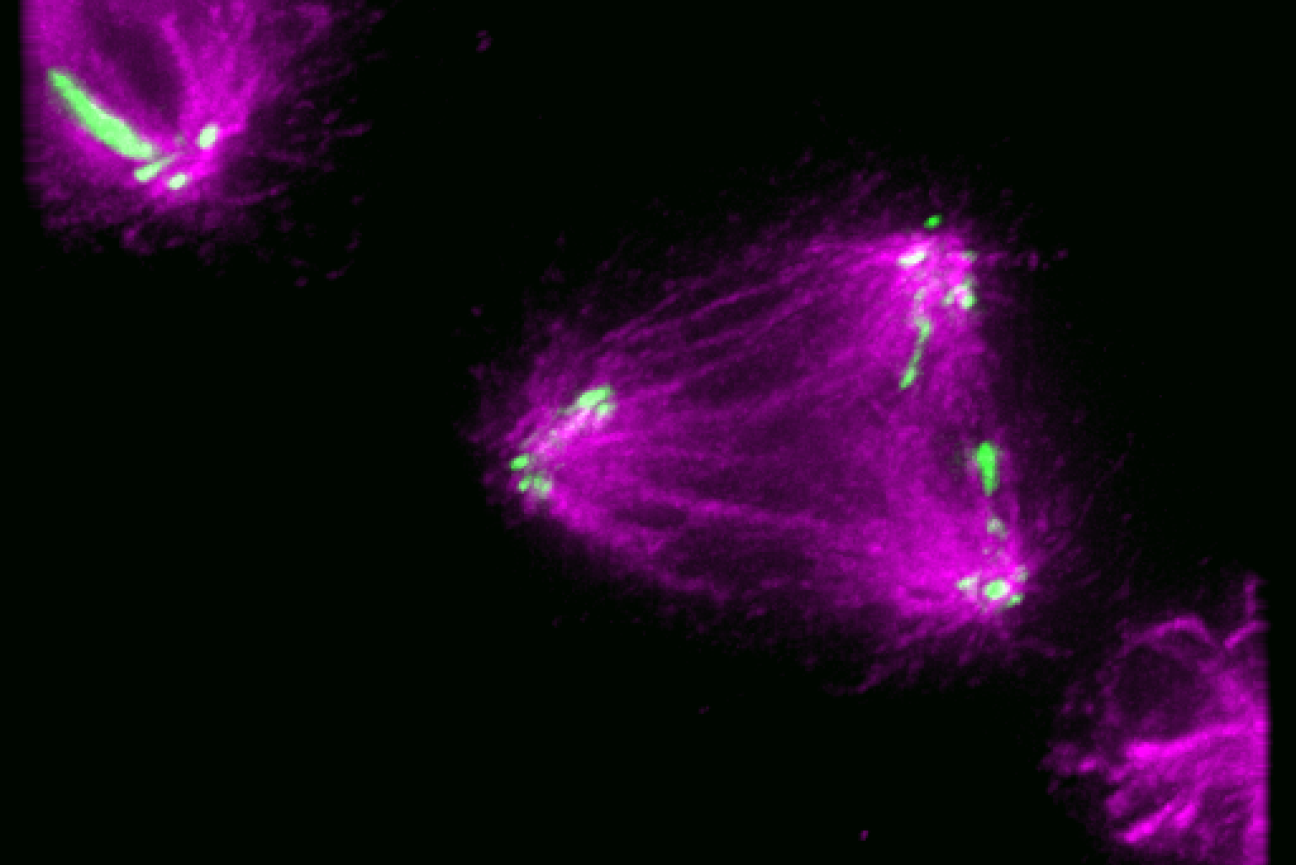
Kinetochore-like CENP-T particles (green) interact with microtubules of a malformed mitotic spindle that has three rather than two poles during chromosome segregation.
Gunter Sissoko
How cells accurately assemble complex machinery
Proteins are the workhorses of the cell, carrying out functions to keep everything running smoothly. Some proteins work on their own, but in other cases many proteins assemble together to create a complex machine. These proteins are able to do more working cooperatively than they could alone, the same way a single motor is powerful but not nearly as useful as a motor combined with other parts to make a car. The instructions for building each individual protein come from DNA, but researchers do not completely understand how cells regulate the assembly of many proteins into a larger machine. Not only must cells assemble their machinery accurately, but they must do so at the correct time and in the correct location for the machinery to perform its task—otherwise, the machine can fail or cause damage as it does its work incorrectly.
One important task for which the cell relies on complex machinery is the division of chromosomes during cell division. When a cell divides, it duplicates its chromosomes and then carefully organizes and distributes them so that each daughter cell ends up with a complete and accurate set of chromosomes. During cell division in humans and many other species, pairs of matching chromosomes line up in the center of the cell. Cellular machinery assembles at a central point on each chromosome called the centromere to separate the two chromosomes in each matching pair and pull them to opposite ends of the cell, where they join what will become the two daughter cells. If the machinery does not assemble exactly at the centromere, it can tear the chromosomes apart or sort them incorrectly into the new cells. These errors can kill the cells or create defects that may contribute to disease.
The machinery that attaches to the centromere and helps to correctly sort and transport the chromosomes is called the kinetochore. In humans, it is a massive complex made up of many copies of many different proteins. Despite the importance of kinetochore location for proper cell division, researchers did not know how cells control where on chromosomes the kinetochore assembles. Whitehead Institute Member Iain Cheeseman, then-graduate student in his lab Gunter Sissoko, Perelman School of Medicine at the University of Pennsylvania Associate Professor Ekaterina Grishchuk, and graduate student in her lab Ekaterina Tarasovetc developed a set of tools that allowed them to solve this mystery, as detailed in the journal Nature Cell Biology on January 2.
The researchers found that the determining factor in where the kinetochore assembles is the local concentration of kinetochore molecules: enough of the molecules need to be near each other in the same space to trigger assembly. The researchers determined this by developing kinetochore-like particles that allowed them to study aspects of how large numbers of kinetochore proteins interact when in close proximity to each other versus when they are far apart.
“We did not understand why the kinetochore has to be a big complex with so many copies of its many components,” Sissoko says. “Now we know that the density of kinetochore proteins that this creates is necessary for assembly of the whole structure.”
CENP-A marks the spot, but can stray
This project began with a puzzling observation. A protein called CENP-A marks the centromere at all times, and forms the very base of the kinetochore: all other parts of the structure will assemble on and around CENP-A. Therefore, one might assume that CENP-A determines the location of kinetochore assembly. However, CENP-A can sometimes be found outside of the centromere. For example, in cancer cells or in cells where researchers artificially increase CENP-A production, the protein may embed itself elsewhere on the arms of the chromosome, and yet kinetochores do not assemble on top of these errant CENP-A molecules. It made sense that if CENP-A was prone to leaking outside of the centromere, the cell must have some other mechanism to prevent aberrant kinetochore assembly, as the results could be disastrous for the cell—but what was the mechanism?
The researchers suspected that kinetochores might only assemble at a location that had a high concentration of CENP-A, like the centromere, and not at locations that had only a little bit of CENP-A. Other processes in the cell are known to be regulated in a similar fashion: molecules get concentrated in the same small space to facilitate their interactions. CENP-A is hard to study because it is embedded into the chromosome, so to test this hypothesis, the researchers decided to look at CENP-T, a protein that is part of the same kinetochore substructure as CENP-A. Together, many copies of that substructure form the inner kinetochore, which serves to anchor the complex to the centromere and then recruit or trigger the assembly of the outer kinetochore. CENP-T plays a critical role in recruiting the outer kinetochore, which connects chromosomes to microtubules, the cell’s highway system that is used to pull the chromosomes apart.
The researchers created what were essentially large balls of CENP-T and other connective molecules that would not interfere with their function. These balls recreated the density of CENP-T that would be found in a kinetochore. They also created another conglomerate of CENP-T in which they could precisely control the number of CENP-T molecules in the group and then measure how different size groups affected the whole’s ability to recruit outer kinetochore proteins.
“Working together, our labs established a novel experimental system to recreate human kinetochore particles,” Tarasovetc says. “Not only has this allowed us to explore how cells control the formation of functional kinetochores at specific times and locations, but the particles also serve as excellent tools for studying other questions of interest, such as the mechanisms of chromosome motion.”
Using these tools, the researchers found that CENP-T was much better at binding outer kinetochore proteins when surrounded by other CENP-T molecules than when one CENP-T was working alone—and likewise, that bigger groups of CENP-T were better than small groups. Each CENP-T molecule is able to directly bind two molecules of NDC80, a critical component of the outer kinetochore. When the researchers looked at CENP-T in a large group, on average every CENP-T had bound the maximum number of NDC80 molecules. However, when they looked at CENP-T molecules working alone, most of the individual CENP-T molecules had failed to bind even one NDC80.
“There’s a regulatory switch that flips when the inner kinetochore recruits enough CENP-T, that allows CENP-T to recruit the outer kinetochore,” says Cheeseman, who is also a professor of biology at the Massachusetts Institute of Technology. “When you have that protein by itself, it still has all those binding interfaces but it isn’t using them. When you reach that threshold density of CENP-T, suddenly it can really seed formation of these structures.”
The particles that the researchers created function so similarly to human kinetochores that the researchers intend to use them to answer more questions about kinetochore function. They hope that other researchers will likewise make use of their approach to study the kinetochore or, more broadly, investigate how the local concentration of different proteins affects their function. The researchers are also working on figuring out the mechanism by which CENP-T becomes better at binding NDC80 when surrounded by other CENP-T molecules.
“While it's common knowledge that kinetochores assemble due to the binding of proteins like NDC80 and CENP-T in a specific sequence, our study revealed a delightful surprise. The process is not as straightforward as it seems, and binding of NDC80 to CENP-T is dependent on whether CENP-T is in a clustered form,” Grishchuk says. “We’re excited to learn more about the underlying molecular mechanism.”
Citation:
Gunter B. Sissoko, Ekaterina V. Tarasovetc, Océane Marescal, Ekaterina L. Grishchuk, and Iain M. Cheeseman. "Higher-order protein assembly controls kinetochore formation." Nature Cell Biology (2024). https://doi.org/10.1038/s41556-023-01313-7
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu


