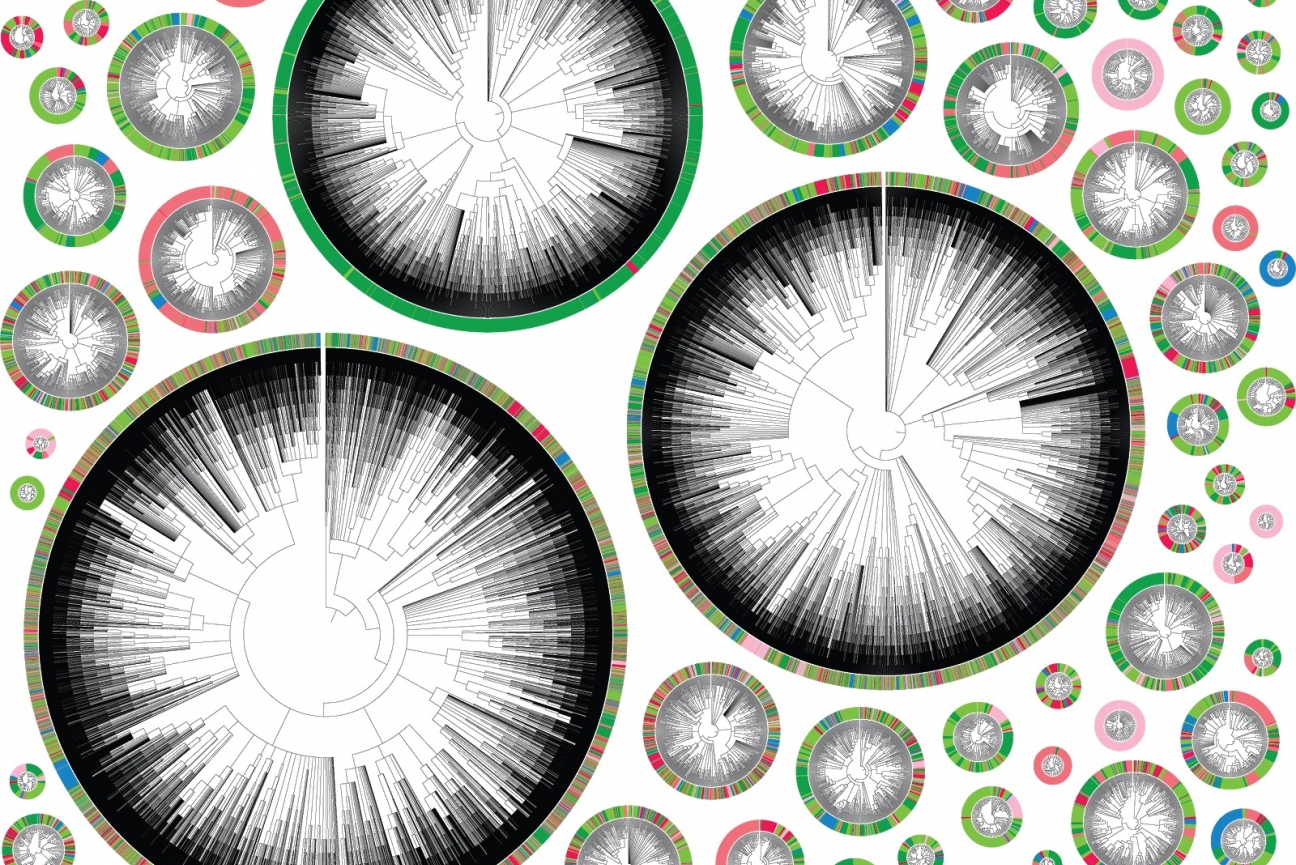
Phylogenetic trees starting with an individual cancer cell. Each color represents a different location in the body. A very colorful tree shows a highly metastatic phenotype, where a cell’s descendants jumped many times between different tissues. A tree that is primarily one color represents a less metastatic cell.
Jeffrey Quinn/Whitehead Institute
Catching cancer in the act
When cancer is confined to one spot in the body, doctors can often treat it with surgery or other therapies. Much of the mortality associated with cancer, however, is due to its tendency to metastasize, sending out seeds of itself that may take root throughout the body. The exact moment of metastasis is fleeting, lost in the millions of divisions that take place in a tumor. “These events are typically impossible to monitor in real time,” said Whitehead Institute Member Jonathan Weissman.
Now, researchers led by Weissman, who is also a professor of biology at Massachusetts Institute of Technology and an investigator with the Howard Hughes Medical Institute, have turned a CRISPR tool into a way to do just that. In a paper published January 21 in Science, Weissman’s lab, in collaboration with Nir Yosef, a computer scientist at the University of California, Berkeley, and Trever Bivona, a cancer biologist at the University of California, San Francisco (UCSF), treats cancer cells the way evolutionary biologists might look at species, mapping out an intricately detailed family tree. By examining the branches, they can track the cell’s lineage to find when a single tumor cell went rogue, spreading its progeny to the rest of the body.
“With this method, you can ask questions like, ‘How frequently is this tumor metastasizing? Where did the metastases come from? Where do they go?’” Weissman said. “By being able to follow the history of the tumor in vivo, you reveal differences in the biology of the tumor that were otherwise invisible.”
Scratch paper cells
Scientists have tracked the lineages of cancer cells in the past by comparing shared mutations and other variations in their DNA blueprints. These methods, however, depend to a certain extent on there being enough naturally occurring mutations or other markers to accurately show relationships between cells.
That’s where Weissman and co-first authors Jeffrey Quinn, then a postdoctoral researcher in Weissman’s lab, and Matthew Jones, a graduate student in Weissman’s lab, saw an opportunity to use CRISPR technology — specifically, a method developed by Weissman Lab member Michelle Chan to track embryo development — to facilitate tracking.
Instead of simply hoping that a cancer lineage contained enough lineage-specific markers to track, the researchers decided to use Chan’s method to add in markers themselves. “Basically, the idea is to engineer a cell that has a genomic scratchpad of DNA, that then can be ‘written’ on using CRISPR,” Weissman said. This ‘writing’ in the genome is done in such a way that it becomes heritable, meaning a cell’s grand-offspring would have the ‘writing’ of its parent cells and grandparent cells recorded in its genome.
To create these special “scratchpad” cells, Weissman engineered human cancer cells with added genes: one for the bacterial protein Cas9 — the famed “molecular scissors” used in CRISPR genome editing methods — others for glowing proteins for microscopy, and a few sequences that would serve as targets for the CRISPR technology.
They then implanted thousands of the modified human cancer cells into mice, mimicking a lung tumor (a model developed by collaborator Bivona). Mice with human lung tumors often exhibit aggressive metastases, so the researchers reasoned they would provide a good model for tracking cancer progression in real time.
As the cells began to divide, Cas9 made small cuts at these target sites. When the cell repaired the cuts, it patched in or deleted a few random nucleotides, leading to a unique repair sequence called an indel. This cutting and repairing happened randomly in nearly every generation, creating a map of cell divisions that Weissman and the team could then track using special computer models that they created by working with Yosef, a computer scientist.
Revealing the invisible
Tracking cells this way yielded some interesting results. For one thing, individual tumor cells were much different from each other than the researchers expected. The cells the researchers used were from an established human lung cancer cell line called A549. “You’d think they would be relatively homogeneous,” Weissman said. “But in fact, we saw dramatic differences in the propensity of different tumors to metastasize — even in the same mouse. Some had a very small number of metastatic events, and others were really rapidly jumping around.”
To find out where this heterogeneity was coming from, the team implanted two clones of the same cell in different mice. As the cells proliferated, the researchers found that their descendents metastasized at a remarkably similar rate. This was not the case with the offspring of different cells from the same cell line — the original cells had apparently evolved different metastatic potentials as the cell line was maintained over many generations.
The scientists next wondered what genes were responsible for this variability between cancer cells from the same cell line. So they began to look for genes that were expressed differently between nonmetastatic, weakly metastatic and highly metastatic tumors.
Many genes stood out, some of which were previously known to be associated with metastasis — although it was not clear whether they were driving the metastasis or simply a side effect of it. One of them, the gene that codes for the protein Keratin 17, is much more strongly expressed in low metastatic tumors than in highly metastatic tumors. “When we knocked down or overexpressed Keratin 17, we showed that this gene was actually controlling the tumors’ invasiveness,” Weissman said.
Being able to identify metastasis-associated genes this way could help researchers answer questions about how tumors evolve and adapt. “It’s an entirely new way to look at the behavior and evolution of a tumor,” Weissman said. “We think it can be applied to many different problems in cancer biology.”
Where did you come from, where did you go?
Weissman’s CRISPR method also allowed the researchers to track with more detail where metastasizing cells went in the body, and when. For example, the progeny of one implanted cancer cell underwent metastasis five separate times, spreading each time from the left lung to other tissues such as the right lung and liver. Other cells made a jump to a different area, and then metastasized again from there.
These movements can be mapped neatly in phylogenetic trees (see image), where each color represents a different location in the body. A very colorful tree shows a highly metastatic phenotype, where a cell’s descendents jumped many times between different tissues. A tree that is primarily one color represents a less metastatic cell.
Mapping tumor progression in this way allowed Weissman and his team to make a few interesting observations about the mechanics of metastasis. For example, some clones seeded in a textbook way, traveling from the left lung, where they started, to distinct areas of the body. Others seeded more erratically, moving first to other tissues before metastasizing again from there.
One such tissue, the mediastinal lymph tissue which sits between the lungs, appears to be a hub of sorts, said co-first author Jeffrey Quinn. “It serves as a waystation that connects the cancer cells to all of this fertile ground that they can then go and colonize,” he said.
Therapeutically, the discovery of metastasis “hubs'' like this could be extremely useful. “If you focus cancer therapies on those places, you could then slow down metastasis or prevent it in the first place,” Weissman said.
In the future, Weissman hopes to move beyond simply observing the cells and begin to predict their behavior. “It’s like with Newtonian mechanics — if you know the velocity and position and all the forces acting on a ball, you can figure out where the ball is going to go at any time in the future,” Weissman said. “We're hoping to do the same thing with cells. We want to construct essentially a function of what is driving differentiation of a tumor, and then be able to measure where they are at any given time, and predict where they're going to be in the future.”
The researchers are optimistic that being able to track the family trees of individual cells in real time will prove useful in other settings as well. “I think that it's going to unlock a whole new dimension to what we think about as a measurable quantity in biology,” said co-first author Matthew Jones. “That's what's really cool about this field in general is that we're redefining what's invisible and what is visible.”
Jeffrey J. Quinn, Matthew G. Jones, Ross A. Okimoto, Shigeki Nanjo, Michelle M. Chan, Nir Yosef, Trever G. Bivona, Jonathan S. Weissman. “Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts.” Science, Jan. 21, 2021.
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu