If cells could talk
This story is part of our series, Tool and Method Development at Whitehead Institute. Click here to see all stories in this collection.
Cells are constantly sending and receiving information to keep the body running smoothly. Researchers at Whitehead Institute are finding new ways to listen in on these communications, or even to tap into cells’ biology and induce them to “report” to researchers about what is going on in the body.
Read on to learn to hear from Institute Members Pulin Li, a synthetic biologist who has recreated cell-cell communication systems from the bottom up, because the best way to learn how something works is often to take it apart and remake it yourself; Rudolf Jaenisch, who has engineered cells to relay to researchers how much of a certain gene is being produced at any given moment; and Jonathan Weissman, who has designed a technique that embeds “scratchpads” in the DNA of cancer cells, which scientists can use to track the lineage of the cells as they proliferate and metastasize in real time.
Recreating cell social networks in a dish
As a multicellular organism develops, proper tissue patterning depends on the organism’s cells being able to send information to each other. Whitehead Institute Member Pulin Li studies how growing tissues in embryos communicate via concentrations of molecules called morphogens. In her postdoctoral work, she developed a method for recreating these communication systems in the lab.
“I’m interested in really understanding the rules of cell-cell communication and what controls, for example, how far cells can send the information to each other and how they interpret that information,” Li said. “But in a developing embryo, you don't have access to all these cell types and the dynamics of the process.”
Video: cells engineered to send and receive signals, with senders/receivers having different fluorescent colors
So Li developed a way to create synthetic signaling systems, starting on the level of individual cells. She first engineered cells to be either signaling or receiving cells for various molecular signals. “We build fluorescent signal reporters into the cells, so then the cells would glow when they receive the signal,” she said. “Then, we can take movies over a two- or three-day period of time and see the traveling of the signal from one side of the dish to the other side of the dish.”
Finally, Li analyzes these videos with image-analysis algorithms to make sense of the signals.
Li’s morphogen gradients in a dish are an example of reconstitution, in which researchers try to create biological systems from scratch to learn how they work. “In the past, when we study complex biological questions like embryo development, it’s always a top-down approach where you take an embryo and perturb it, mutate a gene and see what happens and how we can break something down,” Li said. “Now, there's a lot more interest about how we can build these complex processes from the bottom up.”
In the future, Li hopes to use her cell communication methods to create organoids, masses of cells grown in the lab that resemble organs, that work more like actual organs in the human body. “If we can accomplish that, I think it would open up a lot more avenues for both understanding the basic science and also for drug development,” she said.
Specially engineered cells give real-time updates to researchers
While Li creates cell communication networks from scratch, two other Whitehead Institute researchers are modifying cells in existing systems to “report” back to researchers about what’s going on around and inside them.
Whitehead Institute Member Rudolf Jaenisch is an expert in stem cells. In the past, Jaenisch has used these pluripotent cells to generate numerous cell types and to create organoids, as well as chimeric mice with human cells that may model cancer better than cells in a dish. In the late 2010s, he turned his toolset to a condition called Rett syndrome.
Rett syndrome occurs when a genetic mutation disrupts a gene called MECP2, a syndrome that affects young girls, causing them to lose coordination, speech, and use of their hands. There is currently no cure for the condition.
In Rett patients KCC2 encoding a transporter protein, which is also implicated in a variety of brain disorders including epilepsy and schizophrenia, is significantly downregulated. Using CRISPR-Cas9 gene editing techniques and stem cell technologies, Jaenisch and collaborators engineered human reporter neurons, which allowed screening for small molecules that could restore normal levels of KCC2 expression.
With this technology in hand, the researchers were able to screen over 900 chemical compounds to see whether they had any effect on the activity of the KCC2 protein in the cells. Out of those 900 compounds, the researchers found 30 that boost the protein production activity of the KCC2 gene and improve symptoms of Rett syndrome. Twenty of those are drugs undergoing clinical trials or have already been approved by the FDA.
And the reporter cells can be engineered to provide information on different proteins as well, not just KCC2. “[The method] can also be applied to many important disease-related genes in the brain,” Jaenisch said.
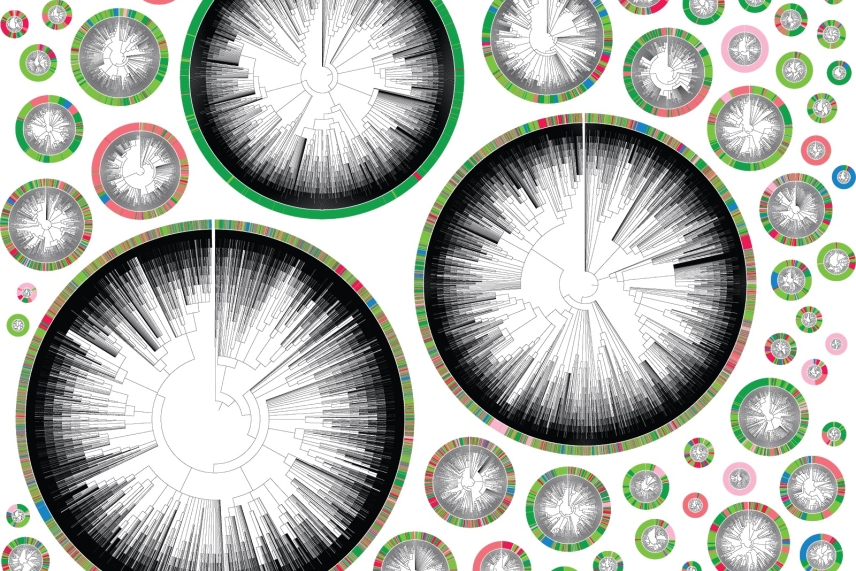
Phylogenetic trees starting with an individual cancer cell. Each color represents a different location in the body. A very colorful tree shows a highly metastatic phenotype, where a cell’s descendants jumped many times between different tissues. A tree that is primarily one color represents a less metastatic cell.
Jeffrey Quinn/Whitehead Institute
Whitehead Institute Member Jonathan Weissman has engineered cells to provide researchers with a different kind of information: instead of reporting on the amount of a particular gene product, Weissman has created cells that record their family history in their DNA. Researchers can then “read” this history and use it to track instances of metastasis in cancer — the moment when cancer cells jump from a primary tumor to a secondary location in the body.
“Every cell comes from the division of another cell, and that means that any set of cells has a family tree that you could reconstruct,” said Weissman. “If you are able to reconstruct these trees, it lets you look at rare events that happened in the past.”
To create a “lineage tracing” system for mapping family trees, Weissman and his collaborators took cells from a cancer cell line — as well as some normal cells — and altered their DNA to contain the necessary materials for CRISPR Cas9 genome editing, as well as a section that basically works like a notepad that can be “written on” by CRISPR. With each generation, the existing writing would be passed down, and a cell several generations down the line would have writing from its parent cells, grandparent cells, and so on.
Reading back the writing on the notepad provides a huge amount of data on which cells are related, which researchers can then analyze with special algorithms to create phylogenetic family trees for each cell that can show where the cancer has traveled throughout the body. (You can read more about the computational side of the method here).
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu


