Of peas and patterns
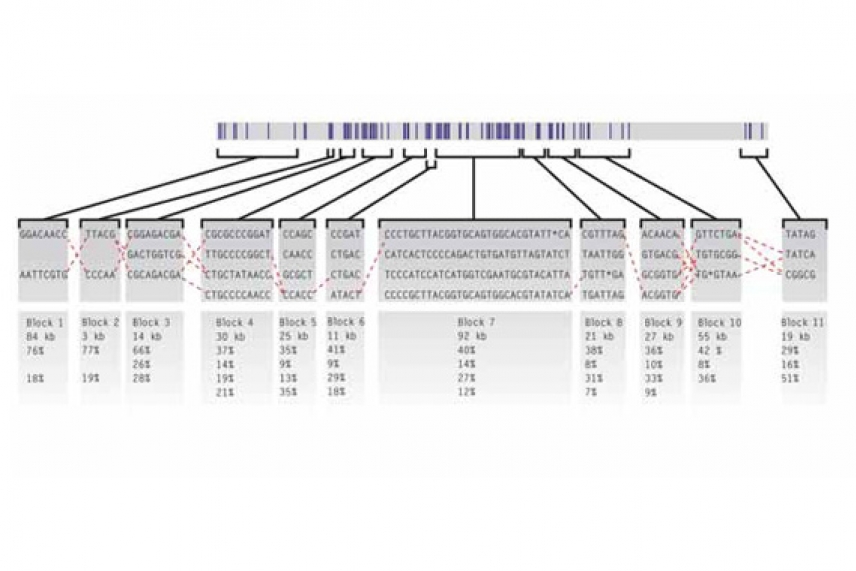
A haplotype map of a portion of chromosome 5 that includes a gene involved in Crohn's disease. Each of the 11 haplotype blocks comes in two to four "flavors," represented by unique combinations of single nucleotide polymorphisms—places in the genetic code where DNA differs from one person to the next by a single letter. The dashed lines show common relationships between the blocks. The percentages indicate the occurrence of each of the common flavors found in the patients studied.
In the 19th century, mathematical formulas didn’t figure much into biology. But when Austrian monk Gregor Mendel crossed and counted his round and wrinkled peas, he found something unexpected: a pattern. His studies showed traits pass from parent to offspring in a predictable fashion, following well-understood rules of mathematics.
Carefully transferring pollen from flower to flower, he bred thousands of pea plants to study the patterns that appeared in succeeding generations. Round or wrinkled, green or yellow, short or tall. From these garden variety traits Mendel learned that pairs of characteristics organize and combine themselves in specific and predictable ways. His study, sadly ignored for years, helped establish the laws of heredity and, ultimately, the field of genetics. It also changed the way biologists approached their work. Mathematics and quantifiable measurement became part of the equation in biological studies.
Today, scientists poring over the human genome catalog are using mathematical and statistical analyses to discover additional patterns of genetic variability. What Mendel did with pencil, paper, and patience now is done with computers and sophisticated mathematical formulas. The studies are revealing combinations that can contribute to disease and point the way to new treatments.
Three years ago, researchers at the Whitehead/MIT Center for Genome Research were working on just such a study when they discovered groups of genes that travel together in the human genome in large, tidy units called “haplotype” blocks. The find was uncovered when DNA analysis expert Mark Daly found a pattern.
A genetic inheritance
It was early 2001, and the recently completed Human Genome Project had, for the first time, made it possible for scientists to compare different parts of the genome, the catalog of chemical units called bases that spell out the genetic code. It’s a catalog written in an alphabet of Cs, Ts, As, and Gs—the letters signifying the bases—and divided into 23 volumes, one for each chromosome in a cell’s nucleus. Sitting in his office at Whitehead Institute, Daly was settling in for a good read.
“It was at a time when the expectation was that these data were going to be very complicated, and that there was going to be no structure or recognizable patterns that we could take advantage of,” says Daly, who is a Whitehead Fellow. “So it was really just a matter of taking an unfettered look at the data, not looking for a particular answer, but simply looking at it for what it was.”
Reading through a series of base pairs, he found that over long stretches of DNA—say 50,000 letters—only a few common genetic variations arose. Scores of people shared the same series of letters across long sequences, as though their genetic inheritance had been handed down in large, prepackaged chunks.
Working with his colleagues in the genome center, Daly further analyzed the blocks using sophisticated analysis techniques. The scientists ended up hypothesizing that these long segments of the genome passed from generation to generation undisturbed by recombination. The group, which included Whitehead Member Eric Lander and research scientist John Rioux, published their findings in Nature Genetics in October 2001.
Additionally, the researchers made a case for using haplotype patterns to study disease, identifying a variant that could put people at high risk for Crohn’s disease, a chronic inflammatory bowel disorder.
Daly then collaborated with Whitehead Affiliate Member David Altshuler to see if haplotype blocks occurred throughout the human genome. “The limitation of that [first] study was that it was just one region of the genome, and it was just one population—a European population,” says Altshuler, who now serves as a founding member of the Broad Institute, a research collaboration headed by Lander that was launched in 2003 by Whitehead, Massachusetts Institute of Technology, and Harvard University and its affiliated teaching hospitals. “Also, it was a disease gene, so it was possible there was something unusual about this region because it caused disease.”
The researchers analyzed 50 different regions of the human genome in samples from Africa, Europe, and Asia. Their findings, published in Science in June 2002, showed that haplotype patterns do indeed appear throughout the entire genome.
Creating a new map
The highly conserved segments of DNA the team uncovered provide an efficient way to wade through the enormous amount of data produced by the Human Genome Project, Daly observes. The findings also helped serve as an impetus for building a haplotype map of the human genome—called HapMap—to describe the common patterns of variation that are found in DNA.
“The enthusiasm for [HapMap] was really sparked by Mark’s observation and his work with collaborators,” Altshuler says. “No one told him to go find this pattern. He looked at the data, he saw what he saw, and he described it clearly. And everyone went and looked and found it in their data, too.”
Launched in the fall of 2002, the HapMap project will allow scientists to more rapidly identify the links between genetic variation and complex diseases, such as diabetes, arthritis, cancer, stroke, heart disease, and asthma. These illnesses can result from an unfortunate conflux of genes and environmental factors, such as diet, smoking, and lack of exercise. Scientists have had difficulty pinpointing the molecular underpinnings of these diseases because, in most cases, multiple genes are at play.
But haplotype mapping may change that. The power of the haplotype pattern lies in its ability to correlate places in the genetic code where DNA differs from one person to the next by a single letter. Called single nucleotide polymorphisms, or SNPs, these tiny changes occur about once in every 1,000 base pairs in the genome, transposing a C to a T or an A to a G.
For the most part—99.7 percent—your genetic blueprint reads just like everyone else’s. But the differences in your code and that of your neighbor are almost all found in these single molecular flips.
What’s more, scientists learned that though a single SNP may have only a subtle effect on a gene or its encoded protein, that small influence can make a person more susceptible to disease, or influence her response to environmental factors and therapeutic drugs.
Looking for patterns
While everything they’d learned to this point suggested the researchers were going in the right direction, the road ahead looked long. Just how were they supposed to decipher that .3 percent of genetic code that makes individuals, well, individual?
Then, Daly uncovered a pattern. When SNPs do occur, he learned, they tend to do so in a predictable fashion, making it possible to predict the identity of dozens of neighboring SNPs. Common patterns emerged, so for any particular gene region, only a handful of common SNP variants, or haplotype patterns, exist. This means that instead of searching base-by-base through all the differences in a particular region of the genome to find one responsible for a disease, researchers may examine a smattering of key SNPs rapidly in large populations.
HapMap researchers in Canada, Japan, the United Kingdom, China, Nigeria, and the United States now are rounding up DNA samples from local families to find genes that affect health, disease, and responses to drugs and other factors. For each genetic variation pattern, scientists eventually will tally the numbers to see how many people carry that version, and of those, how many get the disease and how many don’t.
In developing the haplotype map, scientists also are learning more about how genes organize and sort themselves out to create genetic variation.
Take recombination, for example, the scrambling process used in meiosis to create new genetic recipes.
As cells divide to produce eggs or sperm, chromosome pairs split in half so that daughter cells wind up with only one set of chromosomes. But before separating, the chromosomes swap some of their genetic ingredients so that new genetic combinations are formed. Up until a few years ago, scientists thought recombination was random, and could happen anywhere in the human genome.
Daly’s 2001 study that described haplotype patterns, along with seminal findings from another group, suggested that, perhaps, recombination wasn’t a random process at all. Since then, studies have shown that recombination in the human genome is, instead, clustered in a small number of recombination “hotspots.”
“One of the ancillary benefits of the haplotype map is it’s also providing us a map of all these hotspots,” Altshuler says, “which is useful both in terms of disease-gene mapping and in understanding basic biology.”
Tools of the trade
As a participant in the analysis group for the HapMap project, Daly is working to develop ways to systematically sort through the information. His talent for sifting through data was recognized soon after he entered Whitehead 18 years ago as a physics undergraduate at MIT. Using his knowledge in computational science and mathematics, Daly has developed numerous analytical tools to help researchers find and understand patterns in all types of data. His Haploview software, for example, allows researchers worldwide to access, visualize, and interpret data made available through the Human Genome Project.
“Most of the computational work that we do is not about discovering and developing new mathematical algorithms and techniques. It’s adopting and modifying a lot of well-established techniques in other areas of science to address specific problems,” Daly says.
Ultimately, the HapMap may help scientists uncover genetic nuances that not only lead to disease, but can provide new clinical insights into subtypes of disease. Scientists are beginning to extract such information, Daly says. “In our Crohn’s disease data we have been able to show that this particular risk factor promotes widespread disease of the gastrointestinal system.” Such findings may change how scientists search for and use information on the genetic foundations of disease, Daly says. “People would say, ‘Because disease is complex, we have to use our clinical knowledge to figure out what the distinct subtypes of that disease are so that we can more efficiently make use of the genetics.’ This work suggests in some cases, that may work in reverse—genetics may help lead us to better clinical classification.”
And some benefits, such as using information from an individual’s genetic profile to predict a therapeutic outcome, might come sooner rather than later.
“We have an expectation that some of the discoveries spawned from HapMap will be along the lines of finding genetic variation to predict one’s response—either positively or adversely—to different drugs used to treat disease,” Daly says.
“Wouldn’t it be nice to know, for your particular form of diabetes, which of the many treatment options may be most effective for you or have the fewest side effects?” he asks.
The HapMap project, scheduled to be completed next year, is producing “reams of data.” Scientists will begin tabulating preliminary results to describe genetic variation this fall.
The real challenges, Daly says, lie ahead in decoding the information to figure out how that variation is involved in complex disease.
“The question is, once we have a HapMap and can characterize genetic variation, what does that enable us to do in medical genetics that we can’t do today?” he suggests. “It’s only a tool, which we need to apply intelligently. And depending on the true complexities of these diseases, we don’t know how much work that’s going to be.”
He’s now looking ahead, working to develop mathematical methodologies to help scientists decode the information from the HapMap. When the project is completed, scientists will be able to leaf through millions of human variations. Hidden in there, somewhere, scientists will surely find a pattern.
Susan Gaidos writes about basic research, health, medicine, and the environment. This article first appeared in the fall 2004 issue of Paradigm magazine.
Topics
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu


