New ways to work in the lab
This story is part of our series, Tool and Method Development at Whitehead Institute. Click here to see all stories in this collection.
Much of biology now takes place in the computational world, but the core research at Whitehead Institute still lies in the lab, where researchers conduct experiments using cells and reagents, interact directly with research organisms, and use a variety of hands-on methods to answer their questions.
Sometimes, Whitehead Institute researchers encounter a question for which the right method does not exist yet. When this occurs, rather than being stymied, they tackle the challenge head-on and look to create a new technique to accomplish their goal. Read on to learn about new lab methods developed at the Institute in recent years that are helping researchers study the behavior of elusive axons, create biofuels from agricultural waste, solve the structure of mystery molecules, and reversibly edit the human genome.
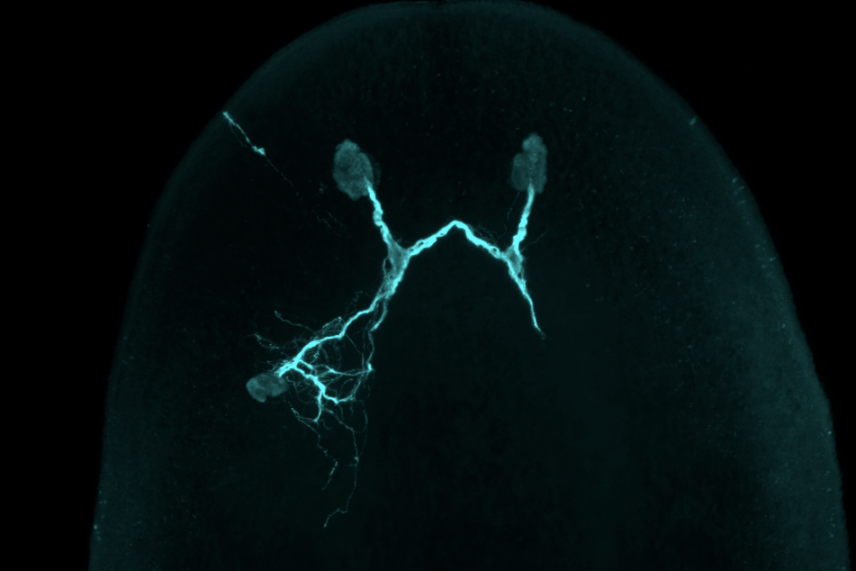
A planarian featuring a transplanted eye.
Kutay Deniz Atabay/Whitehead Institute
An eye for an eye in planarian flatworms
Peter Reddien’s lab at Whitehead Institute studies the science of regeneration using charismatic model organisms called planarians. When these tic-tac-sized flatworms lose one of their cartoonish eyes, a new one soon grows in its place.
In order for an eye to become functional, the cells that make up the eye must be “wired in” to the brain via cellular extensions called axons. When the eye takes in sensory information, the axons convey that information to the brain, where it is interpreted and the animal can respond to it.
The researchers hoped to use this eye regeneration as a window into the targeting of axons in regeneration. “Axon behavior is still a mystery,” said Kutay Deniz Atabay, a postdoctoral researcher in the lab. “We know a lot more now than we knew 50 years ago, but there's still a lot to figure out because biology is noisy. You’re looking at the behavior of this tiny thing — a growing axon — and it's interacting with several molecules around it and making a specific, precise decision.”
In order to study maintenance of ectopic eyes for an earlier project, Atabay and colleagues developed a technique to give a planarian a new eye. The same technique also makes it possible to study growing photoreceptor cell axons in as controlled a situation as possible. “[With this method] you can take an eye from one animal and transplant it into another animal — with or without an eye — and the axonal projections from that eye will correctly wire themselves into the brain,” he said.
But developing the method was no easy feat. “This technique depends on a lot of things going right,” Atabay said. “It actually took us about a year to figure it out. It was a lot of trials, hundreds of experiments — and then one night, one of the transplants worked.”
The method goes like this: First, researchers have to obtain an eye from a donor to transplant into a new animal. This requires a clean, efficient surgery. Then, once they have the eye ready, the researchers prepare the recipient. Usually for planarian surgeries, researchers wash the animals, administer anesthesia, and then begin surgery. But in the case of eye transplants, the researchers realized — by many trials — that the process wouldn’t work without some extra washing/anaesthetizing steps. These extra steps helped induce correct muscle tone needed for the worm’s body to accept the eye. “Basically, the recipient is sitting on a piece of filter paper, and you anaesthetize it, make the initial cut, wash it, anaesthetize it again, and then wash it again,” Atabay said. “And then it's ready to receive the eye.”
Since its development, the eye transplantation method has helped the researchers understand maintenance dynamics of the eyes, interaction of axons with guidepost cells and how certain genes play a role in axon regeneration, and they hope to apply it in different types of experiments in the future. “Having this tool allows us to study the cellular and molecular behaviors of an entire modular organ,” Atabay said. "In this fashion it could be adapted to multiple types of questions."

Whitehead Institute Member Gerald Fink standing in front of a field of the grass Miscanthus giganteus, which is another potential source of cellulose that could be converted to ethanol.
Courtesy of Felix Lam
Lending yeast a helping hand to make biofuels
One method that could diminish greenhouse-gas emissions is to replace petroleum derived fuels with biofuels. For the creation of these cleaner fuels, you have fermentation microorganisms to thank. The lab of Whitehead Institute Founding Member Gerald Fink has developed a method to help these microorganisms create clean fuels from discarded plant matter, opening the door for a new source of renewable energy.
Gasoline today is usually mixed with ethanol, a biofuel made from plant products. In order to create the fuel from the sugars in plants, industrial plants use specially engineered yeasts. The industry currently uses these yeast to ferment the sugar in corn kernels to ethanol, but in the US, there is a huge underutilized source of ethanol: the woody stalks and cobs left over after corn kernels are harvested. These plant materials are full of sugar too — it’s just a little harder to reach and ferment.
That’s because the sugars in these materials are locked up tight in tough molecules called lignocellulose. In order to break down these molecules for the yeast to digest, ethanol producers usually treat the plant materials with an acid wash. This acid bath then creates another problem: toxic byproducts called aldehydes, which kill the yeast required for fermentation. There are ways to make the aldehydes less toxic, but they are not cheap enough for the process to be widely used. And so, that woody corn material is often left in the fields to rot and fertilize the soil.
Enter Whitehead Institute Founding Member Gerald Fink. Fink and collaborators designed a new method to help the yeast along in their fermentation work. The researchers gave the yeast a new gene that helped them convert normally toxic aldehydes into alcohols by sticking on a hydrogen atom. They also supplemented the yeast’s growth media with potassium and adjusting the pH, which helped stabilize the yeast’s cell membranes against high alcohol concentrations. The new method promises a cost-effective way for the ethanol industry to utilize wasted plant materials, creating a cleaner and more efficient fuel industry.
“The combination of these technical advances must be incorporated into modern industrial procedures to increase our ability to use this awesome power of the tiny microbe yeast to produce fuels, vaccines, and chemicals that sustain modern human life,” Fink said.
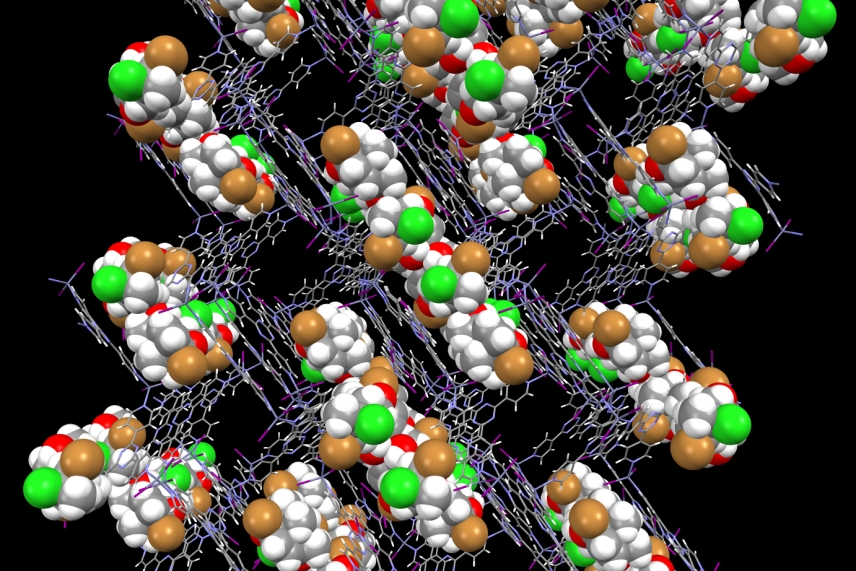
An artistic rendering of molecules held by the crystalline sponge.
Jing-Ke Weng/Whitehead Institute
Crystallizing an idea into a new biochemical method
Plants produce millions of unique small molecules, many of which are unknown to humans. At Whitehead Institute, Member Jing-Ke Weng’s lab is interested in understanding what these products are, how they are made, and how they could be used to inform therapies.
To do this, the researchers in Weng’s lab need to figure out the molecular structure of plant natural products. A tried and true method of understanding molecular structure is to collect a large amount of the molecule, purify it, induce it to form crystals in the lab, and then use a technique called X-ray crystallography to figure out what the molecule looks like on an atomic level.
But for many of the plants Weng studies, it’s difficult to obtain such large amounts of any given natural product — and in fact, such extensive harvesting could be devastating to ecosystems, especially when the plant is rare. “In the old days of plant natural product research, scientists usually went out and collected kilograms of materials,” said Weng. “It's not a very pretty process, and I've heard stories where people went out and essentially cleared out one species of plant over an entire mountaintop.”
That’s why Weng, working in collaboration with Makoto Fujita at the University of Tokyo, has helped develop a method to figure out the molecular structure of plant natural products with a minuscule amount of material. The researchers call the technique the “crystalline sponge method.” The method uses a material made of two complexes of macromolecules which form a lattice — the sponge — that can absorb and envelop molecules of different sizes.
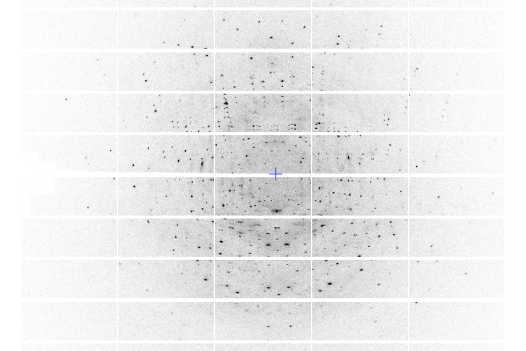
A photo produced by X-ray crystallography that shows the 3D structure of molecules held in the crystalline sponge.
Jing-Ke Weng/Whitehead Institute

Crystalline sponge material
Jing-Ke Weng/Whitehead Institute
“It’s this very unique porous crystalline material,” Weng said. “You can use this material to soak in the small molecule of interest at very low abundance — almost a thousand to a million orders of magnitude smaller than the amount [needed for traditional methods].”
When the molecule of interest is soaked into the crystalline sponge, it’s held static. “Then you can carry on to do standardized X-ray diffraction and solve the structure of the whole complex,” Weng said. “When you do that, you actually solve the structure of the molecule embedded in that material.”
The method has utility beyond the plant sciences as well. “In my view, this is a revolutionary methodology that allows scientists to reveal the unknown structures of very important metabolites,” he said. “The methodology can also be applied very broadly in all fields involving metabolism, not just for plants but also human metabolism for disease biology.”
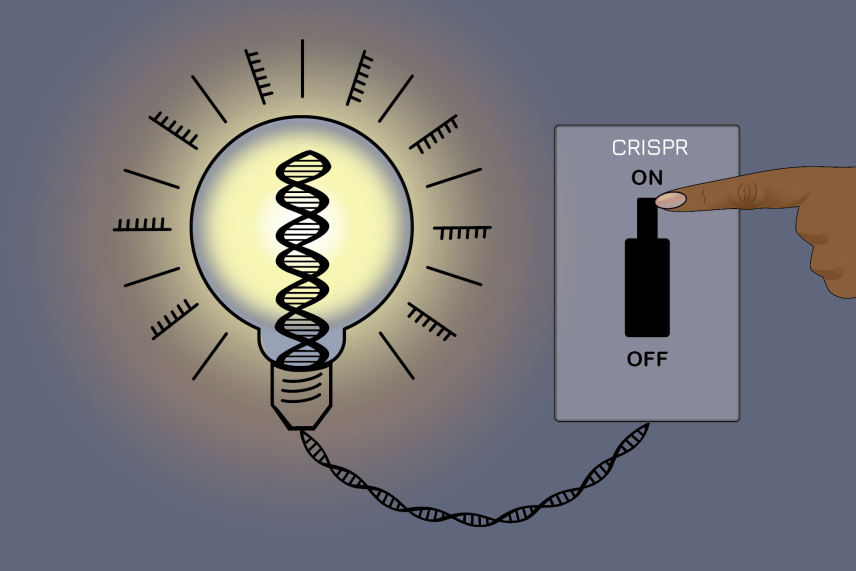
New CRISPR method acts as an on-off switch for gene expression.
Jennifer Cook-Chrysos/Whitehead Institute
Flipping the switch on gene editing
Since its development in 2012, the CRISPR-Cas9 gene editing system has changed the way we study biology. By inducing small breaks in a strand of DNA, researchers can add in or delete genes from the genome, permanently changing the behavior of a cell.
Now, Whitehead Institute Member Jonathan Weissman’s lab has created a new gene editing system: “We have developed an approach we call CRISPRoff that lets you direct CRISPR to a particular gene,” Weissman said. “And instead of changing the DNA by cutting it, which is irreversible and has some toxicity, we can silence the gene [without changing the sequence].”
CRISPRoff can induce changes in gene expression that are passed down through generations, and, unlike the traditional CRISPR-Cas9 method, is fully reversible and does not involve any DNA damage. “This could allow new therapeutic avenues, since we can now turn off genes permanently without having to have any double strand breaks,” Weissman said.
CRISPRoff works by harnessing a natural gene regulation system called methylation, which involves adding small chemical tags called methyl groups onto sections of genes to “silence” them. Methylation — which is the natural system our bodies use to make sure the right genes are expressed at the right time — doesn’t change the sequence of the DNA; it just prevents it from being “read” by cellular machinery.
The researchers created a tiny protein machine that, guided by small RNAs, can tack methyl groups onto specific spots on the strand. These methylated genes are then turned off, hence the name CRISPRoff. Because the method does not alter the sequence of the DNA strand, the researchers can reverse the silencing effect using enzymes that remove methyl groups, a method they called CRISPRon. Weissman anticipates that the method will be useful for conducting fundamental research in the lab, as well as developing therapies for genetic diseases.
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu


