
Unusual Labmates: how C. elegans wormed its way into science stardom
Introduction
Michael Stubna, a graduate student in Whitehead Institute Member David Bartel’s lab, peers into his microscope at the Petri dish full of agar gel below. He spots one of his research specimens, a millimeter-long nematode worm known as Caenorhabditis elegans (C. elegans), slithering across the coating of bacteria–the worm’s food source–on the surface of the gel. The worm leaves sinuous tracks in its wake like a skier slaloming down a slope.
C elegans with tracks clear behind it.
Clip adapted from Video S8 from Edwards S, Charlie N, Milfort M, Brown B, Gravlin C, Knecht J, Miller K (2008). "A Novel Molecular Solution for Ultraviolet Light Detection in Caenorhabditis elegans". PLOS Biology. DOI:10.1371/journal.pbio.0060198. CC BY 2.5.
Michael looks up from the microscope and grabs his worm pick, a metal wire sticking out of a glass tube. He runs the end of the pick through a Bunsen burner flame until the wire glows red, using heat to sanitize the tool. Then he returns his attention to the microscope. He nudges the Petri dish to re-center the worm, and, once the pick has cooled, he coats the tip with some of the sticky bacterial food and uses it to skillfully pluck the worm from the surface of the gel. He puts a fresh dish of agar under the microscope, and presses the pick, with the worm still adhered, to the surface. Almost immediately, the worm sets off, carving fresh tracks into the pristine bacterial lawn.

Michael sanitizes his worm pick before moving C. elegans between plates.
Madeleine Turner/ Whitehead Institute
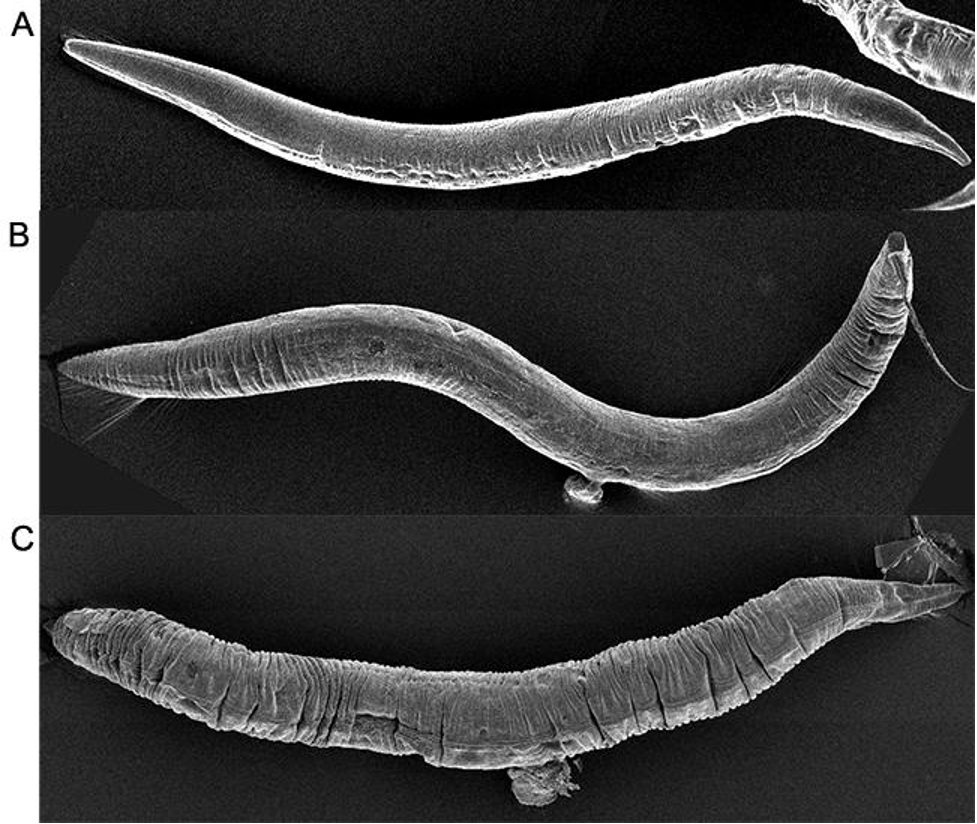
Michael is cultivating C. elegans in order to use them to study microRNAs, tiny RNA molecules involved in gene regulation. Right now, Michael is the only researcher in the Bartel lab using the worms, but in the wider research world, C. elegans is a popular model organism. At first glance, C. elegans is a rather unassuming animal. Barely large enough to see with the naked eye, in nature the worms reside in soil and decomposing vegetation, feasting on bacteria. Except for their heads, their bodies can only bend up and down, so the animals crawl on their sides. The worms have simple tube bodies and are capable of a limited range of behaviors. Nevertheless, researchers frequently turn to C. elegans to learn about not only their biology, but our own. C. elegans is one of the most intimately understood species in biology—the first animal to have its complete genome sequenced or its neural circuitry completely mapped. How did this simple worm become so well studied and a fixture in laboratories around the world?
Making a model
The species C. elegans was first identified, and used in research, after being found in the soil in Algeria around 1900. However, its popularity as a research model skyrocketed in the 1970s, after biologist Sydney Brenner, then at the Medical Research Council Laboratory of Molecular Biology and later the founder of the Molecular Sciences Institute, made the case for it as the best new model species for the field of molecular biology. [1], [2]
What makes C. elegans such a good model organism? The worm exists in a “just right” zone of biological complexity: it is complex enough to have many of the features that researchers want to study, but simple enough that those same features can be examined comprehensively. For example, each C. elegans has 302 neurons, which is enough to be a useful model for everything from questions about how brains form, to how they sense and respond to stimuli, to how neuronal pathways give rise to specific behaviors, to how different diseases cause neurodegeneration. At the same time, 302 neurons is a small enough number for researchers to be able to study each individual neuron and its connections thoroughly. (In comparison, a fruit fly has around 100,000 neurons.)
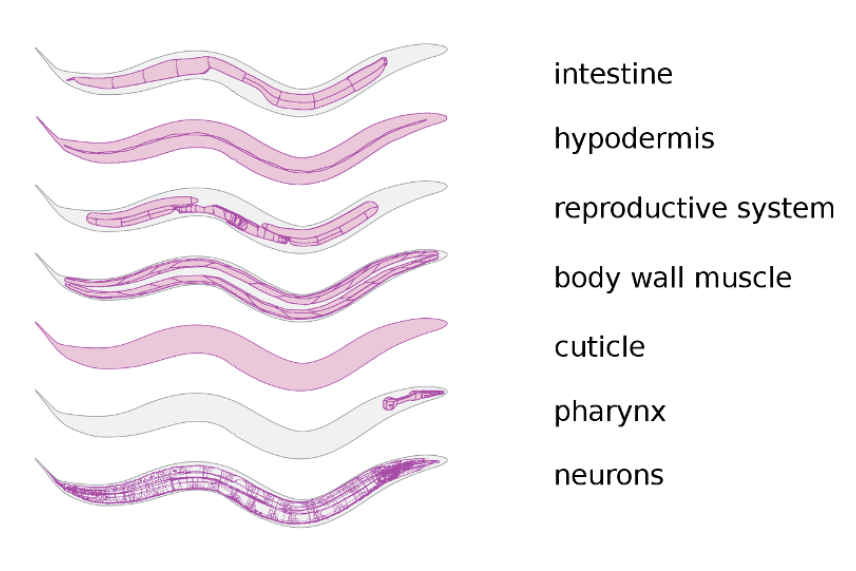
C. elegans has enough anatomical complexity with which to ask interesting questions, yet is simple enough to study in comprehensive detail.
In the same way, C. elegans has just enough complexity to be used to model other common aspects of animal biology, including muscle function, reproduction, digestion, wound healing, aging, and more. It shares many genes with humans and can even be used to model human disease. For example, researchers have used C. elegans to model neurodegenerative diseases such as Alzheimer’s and Parkinson’s.
Additionally, C. elegans has many advantages as a research subject. The worm’s skin is transparent, so researchers can easily observe and capture images of changes occurring inside of its body down to a cellular or even sub-cellular level.
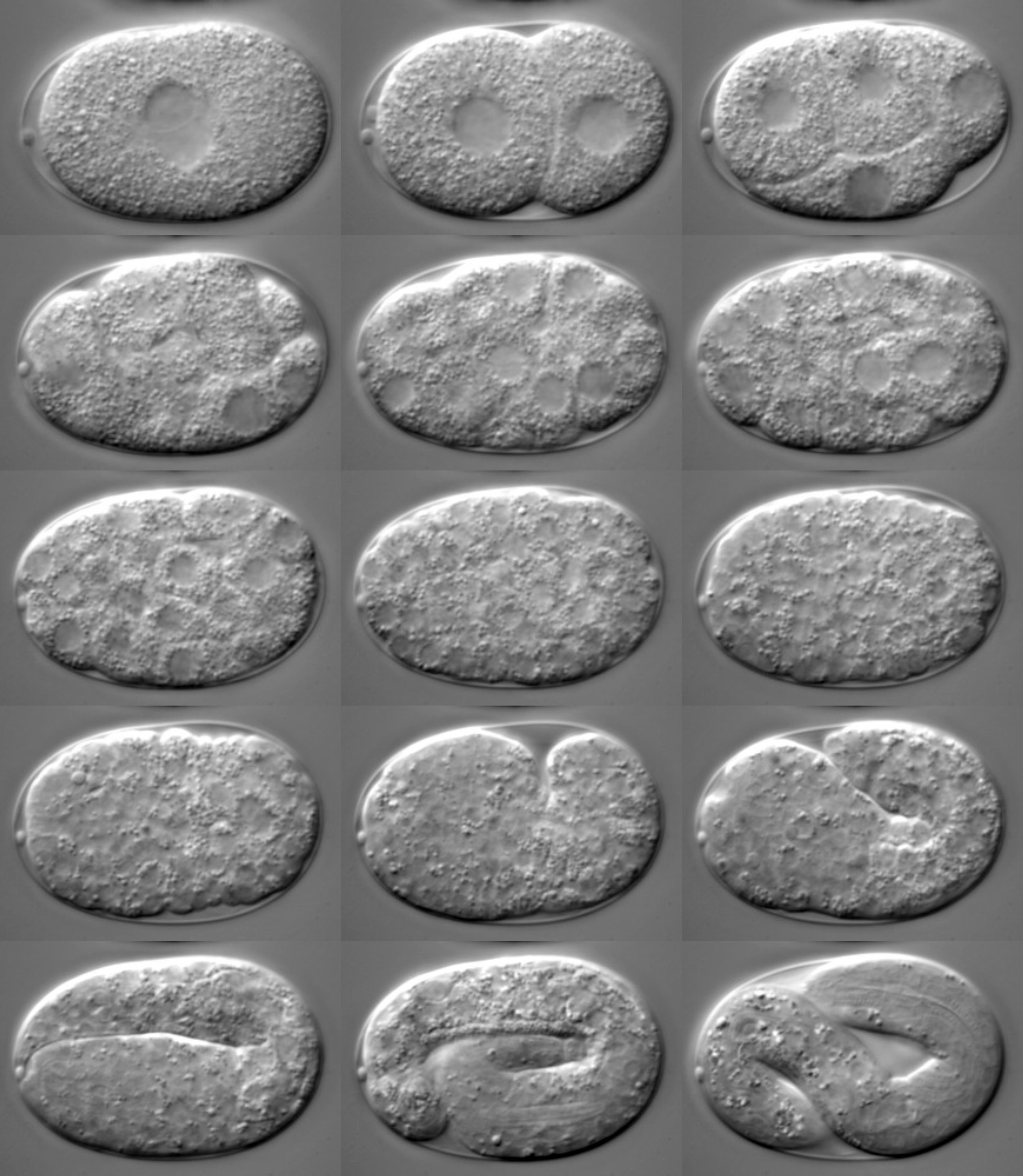
C. elegans is small, hardy, and easy to rear in the laboratory. Fed a simple diet of bacteria and kept at 20 degrees Celsius, C. elegans will mature from an egg, through four larval stages, to a fertile adult in three days. It can then rapidly reproduce to provide researchers with thousands of specimens. C. elegans live for about three weeks, allowing for quick generational turnover, but if researchers want to keep the worms alive for longer, this is easy to do by putting the worms in stasis.
In nature, it is common to find the worms in a state of suspended animation, in which they can survive for months. [3] During poor environmental conditions, such as when food is scarce, instead of maturing into their usual third larval stage, the worms will enter what is known as a dauer stage, a hardier but inactive larval form. When environmental conditions improve, the worms exit the dauer stage and resume normal development. Researchers can recreate this process in the lab.
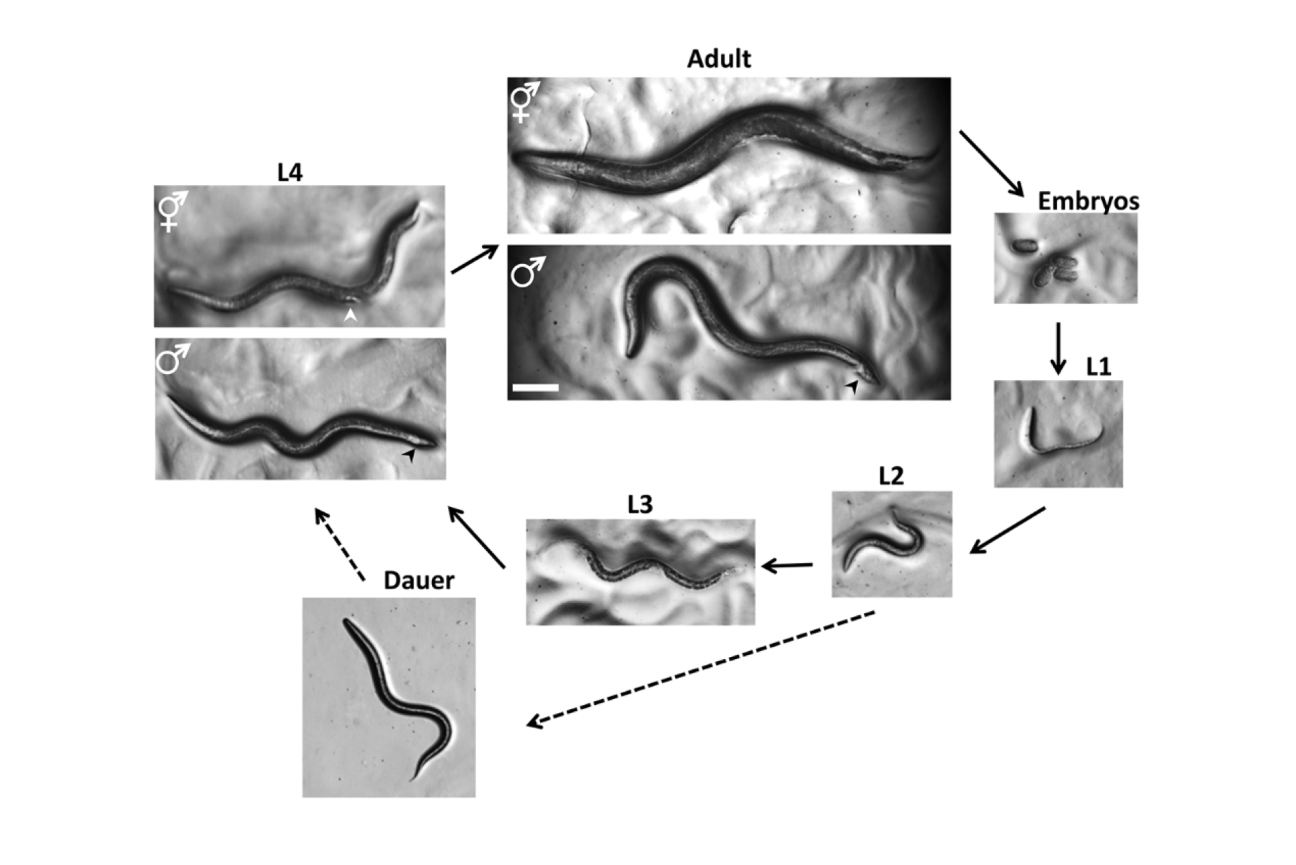
C. elegans’ life cycle. When faced with environmental stress, the worm enters the alternative Dauer stage instead of its usual third larval stage (L3).
“Figure 2. Life Cycle of C. elegans” from Corsi, Ann K., Wightman, Bruce, Chalfie, Martin, “A Transparent window into biology: A primer on Caenorhabditis elegans,” Workbook, ed. The C. elegans Research Community, WormBook, June 18, 2015, 1551-8507, 10.1895/wormbook.1.177.1.
If a researcher is going out of town for a few days, they can keep their worms in a refrigerator at 4 degrees Celsius. They also have a simple solution for storing worms long term: freezing them. Worms put in a negative 80-degree Celsius freezer can survive for years and still be recovered. This makes the worms much easier to maintain than other common model organisms, which need constant maintenance to keep them alive, fed, and reproducing.
“With most model organisms, if you go away on vacation you need to find someone to look after your specimens while you are gone,” Stubna says. “That’s not necessary with worms.”

The fridge where Michael keeps his stock. Credit: Madeleine Turner/ Whitehead Institute
Madeleine Turner/Whitehead Institute
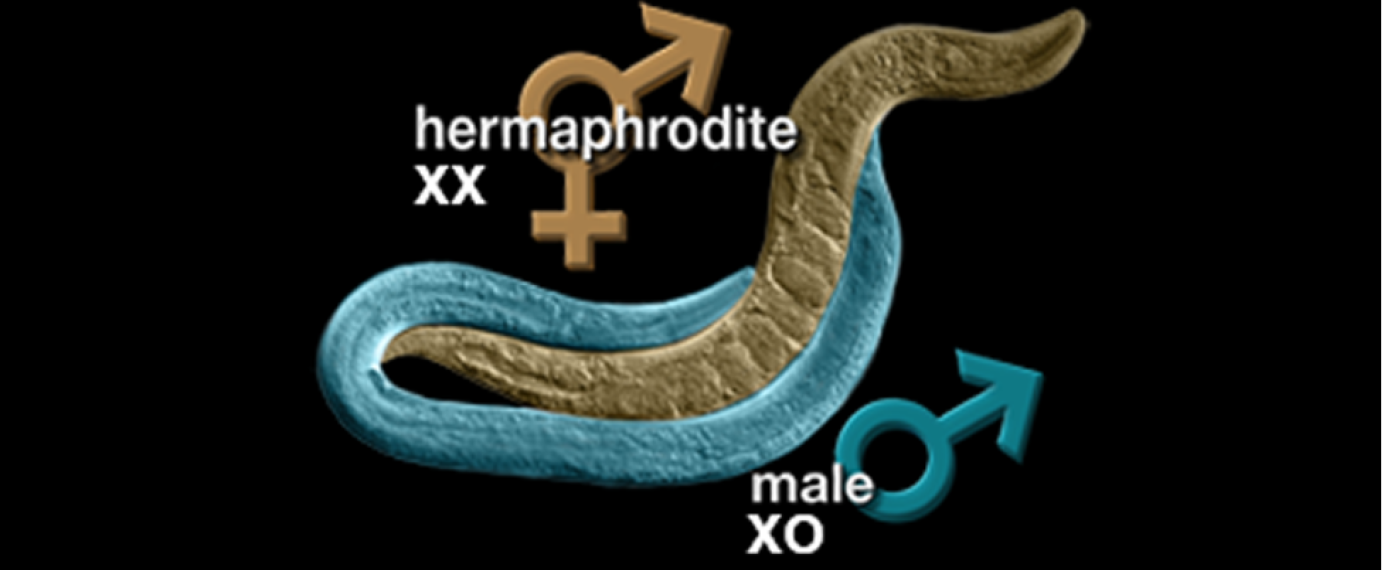
Another advantage of C. elegans is how easy it is to generate large numbers of them. Most C. elegans are self-fertilizing hermaphrodites. They cannot mate with each other, but each worm can generate hundreds of offspring on its own. The self-fertilizing nature of C. elegans generally works in researchers’ favor, as along with making it easy to generate lots of new worms, self-fertilization makes genetic inheritance simpler to predict and manipulate in order to maintain a desired mutation throughout the generations. However, sometimes researchers may want to cross-breed their specimens, for example to combine mutations found in separate adults in an offspring.
Fortunately for researchers, the rare C. elegans worm is male—around .1-.2% in the wild [4] — and males can fertilize hermaphrodites’ eggs, enabling cross-breeding. C. elegans males arise in the wild due to a deviation during sex cell division. Hermaphrodite C. elegans have two X chromosomes. When one of these fails to form correctly, the resulting worm with its one X chromosome will be male, having some key anatomical differences. If researchers need lots of males for their work, they can increase the percentage of a hermaphrodite’s offspring that are male by exposing the worm to heat before it reproduces or by using genetic manipulation. Then, researchers can selectively breed male-heavy populations to further increase the ratio over time.
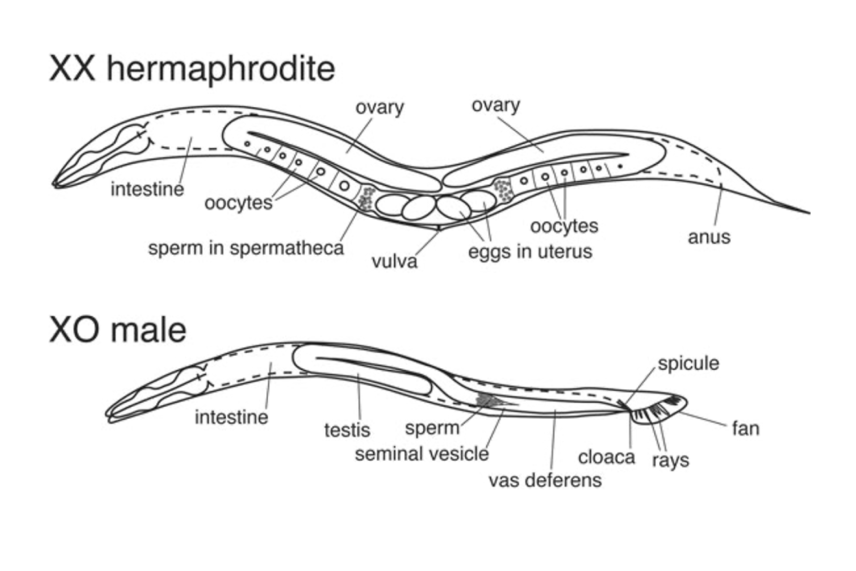
The two sexes have differences in anatomy, including their overall size and their tail structures.
Figure 1 from Zarkower, David, Somatic sex determination (February 10, 2006), Wormbook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.84.1, http://www.wormbook.org.
Researchers have developed a variety of tools and approaches over the years with which to manipulate C. elegans genetically. These have enabled researchers to learn a lot about both the worm’s genes, and genes that it shares with humans and other animals. One useful approach is the development of marker strains.
Getting to know C. elegans inside and out
After Brenner landed on C. elegans as an ideal model, his research group began several ambitious projects to comprehensively understand the worm’s biology. In the following decades, the worm’s anatomy and genome would be detailed in unprecedented detail. The more that researchers learned about the worm, the better a model it became.
Creating a complete cell lineage and neural map
C. elegans is remarkable in that every worm has the same exact number of cells: 959 in the adult hermaphrodite (not counting the cells that will become eggs or sperm). 302 of these cells are neurons. Researchers in Brenner’s group created two first-of-their-kind resources documenting the details of this biology. First, they mapped the worms’ complete cell lineage, recording every cell division that occurs during the worms’ development from fertilized egg to adult. This resource makes it easy for researchers to study how different factors contribute to—or can alter—this development.
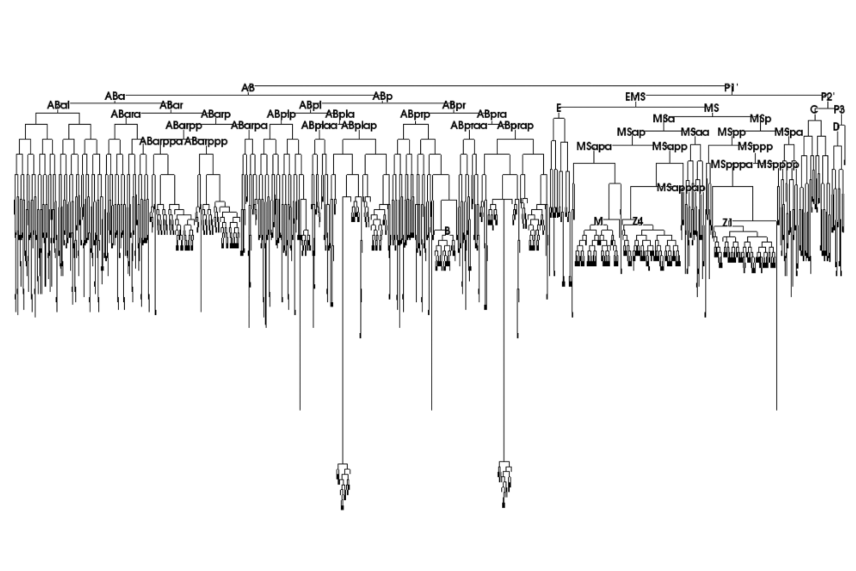
The complete cell lineage of C. elegans. This chart can be used to find the relationships between cells. For example, cell AB divides to form cells ABa and ABp. Researchers can use that information to understand things like the different states that cell lineages go through on their way to creating the adult body’s final cells.
“Complete cell lineage of C elegans” by Andrej Cedilnika, Jeffrey Baumesb, Luis Ibanezb, Sean Megasonc, and Brian Wylied.
Then, the researchers created a wiring diagram, or connectome, of the hermaphrodite worm’s 302 neurons and their thousands of synapses—the junction points where neurons interact. Researchers have used this wiring diagram to identify neurons involved in many different behaviors in C. elegans, as well as to understand how brains form and function across animal species. [5,6] C. elegans was the first, and as of 2021 the only, animal to have had its brain completely mapped.
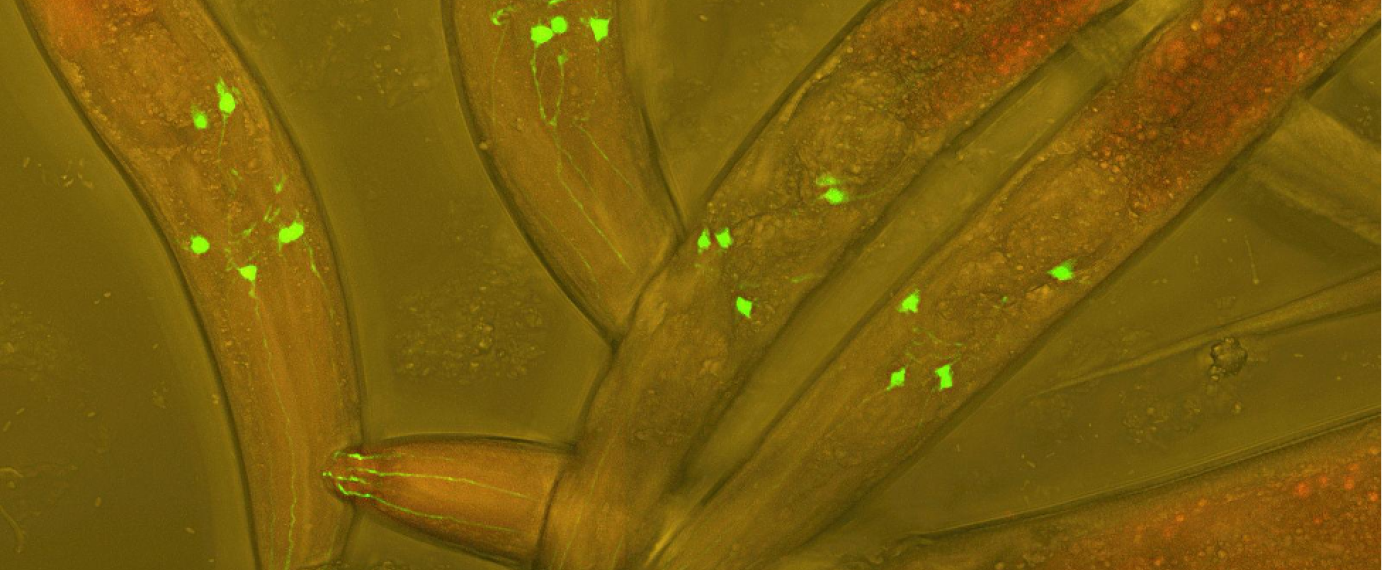
C. elegans with neurons visible in green.
“Varbuss” by Heiti Paves, CC BY-SA 3.0.
Capturing the complete genome
In 1998, C. elegans made the news as the first animal to have its complete genome sequenced. The completion of the 15-year-long sequencing project, helmed by the C. elegans Sequencing Consortium, was announced in a special issue of Science. Researchers had previously compiled complete genomes for a variety of single-celled species, but as an animal, C. elegans had a significantly larger genome. The complete genome provided many useful insights into individual genes, and the relationships between genes both within C. elegans and between species. The ambitious project also proved instructive for how to sequence large genomes. In fact, the Human Genome Project helped to fund the sequencing of C. elegans as a stepping stone to the ultimate goal of sequencing the human genome, which was achieved in 2003.

With C. elegans’ genetics, anatomy, and other biology so thoroughly documented, the worms became an even more potent model organism. Researchers now had a wealth of foundational knowledge about the worm that they could use to make and test hypotheses about specific questions.
Worm culture
As the use of C. elegans in science grew, a community formed among the worm’s researchers. The C. elegans community was quick to develop and share resources. The Worm Breeder’s Gazette is a semi-annual newsletter first published in 1975, which shares information of interest to the C. elegans community such as experimental techniques and new findings. The Caenorhabditis Genetics Center (CGC), founded in 1979, is a central repository from which researchers can order thousands of different strains of C. elegans for use in their own research.

From the early years, prominent researchers working with C. elegans believed strongly in sharing data both among researchers and with the public. This openness set the tone for the field of molecular biology more broadly; for example, open data sharing policies around the sequencing of the C. elegans genome encouraged the Human Genome Project to follow suit. [10]

Several covers of the Worm Breeder's Gazette
The Worm Breeder’s Gazette, http://wbg.wormbook.org/
The worm community often had to build its own tools in order to share data on the scales its members desired. One big project was the creation of ACeDB (A C. elegans Database), a database management system capable of storing and displaying many different kinds of biological information about C. elegans, including its complete genome, in a user-friendly way. The current iteration of ACeDB, known as WormBase, contains the annotated genomes of C. elegans and related nematodes, information on every known C. elegans gene and its function, genetic maps, the C. elegans cell lineage and connectome, and much more. The ACeDB software was soon used to create similar databases for other model organisms. Such databases now exist for many different species, making detailed biological data widely available to everyone. These databases are also often used to share the latest research, maintain a consensus around scientific terminology and gene annotation, and provide educational resources on the model organism. Anyone looking for general information on C. elegans can also visit WormBook, an open access, online review of C. elegans biology.

Many resources on C. elegans are available, both online in the form of databases such as WormBook, WormAtlas, and WormBase, and in person, such as during the annual International C. elegans Conference hosted by the Genetics Society of America.
Clockwise from left: Screenshot of WormBook taken 09/08/2022; “Royce Hall occupied” by Adam Fagen, CC BY-NC-SA 2.0; screenshot of WormBase.org taken 09/06/2022; screenshot of WormAtlas.org taken 09/06/2022.
With these resources and others, the C. elegans community fostered a culture of sharing and scientific openness that continues to this day.
A few of the many discoveries and further tool development
C. elegans research, with its wealth of experimental tools and methods, pre-existing data with which to build and test hypotheses, and a worldwide community happy to share resources, has been the source of many important discoveries over the years. Many of these discoveries have also added to researchers’ toolkits, providing new ways to experiment with C. elegans and other research specimens. A few of these myriad impactful discoveries are highlighted below.
Insights into development and programmed cell death
Brenner and two researchers whom he had mentored, John Sulston, then at the The Wellcome Trust Sanger Institute, and Robert Horvitz, then at the Massachusetts Institute of Technology (MIT), were awarded the first Nobel Prize for work done in C. elegans in 2002. When the researchers were creating the C. elegans cell lineage map, they saw that some cells created during development died off at particular moments, and that this programmed destruction of cells, called apoptosis, was an essential part of creating the adult body. They identified key genes that regulate apoptosis, and their work led to insights into the role of apoptosis in human development, as well as in health and disease. For example, cancer cells are able to avoid apoptosis, and many modern cancer therapies work by reenabling apoptosis of cancer cells.
Andrew Fire at the Stanford University School of Medicine and Craig Mello at the University of Massachusetts Medical School used C. elegans to discover RNA interference (RNAi), a process that cells use to stop genes from being expressed. RNAi became an important research tool after researchers figured out how to tailor RNAi to turn off genes that they are interested in studying in different cells and species. Researchers turn off a gene and see what changes, which helps them figure out the gene’s function. People have also found uses for RNAi in medicine and industry. RNAi is easy to use in C. elegans — researchers can apply it to worms by simply feeding them modified bacteria — so this tool made the worms an even better model for genetics research. Fire and Mellow earned a Nobel Prize for their discovery in 2006.
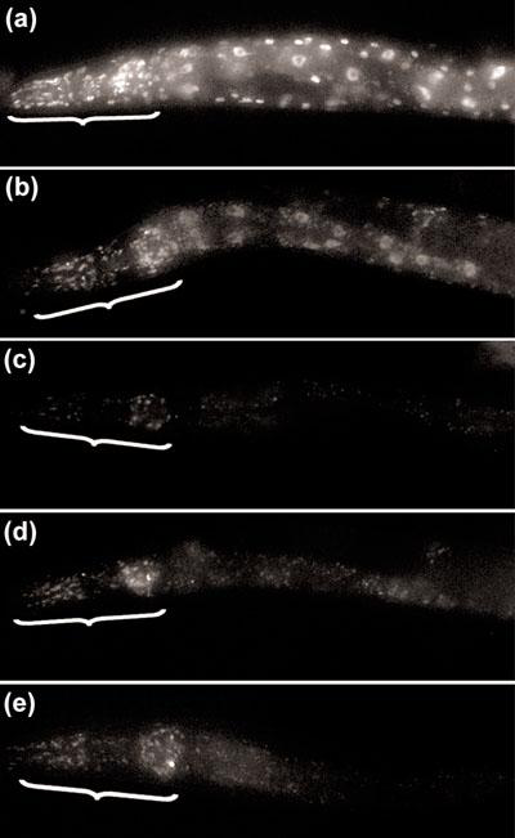
Introducing a new visual tag
C. elegans also contributed to the development of another popular and powerful research tool, green fluorescent protein (GFP). GFP is a protein first found in jellyfish. It glows green under certain light waves. Martin Chalfie at Columbia University showed in C. elegans that the genetic code for GFP could be added as a tag to genes of interest, and then the products of those genes would glow, providing researchers with a great visual marker of where and when the genes were expressed. Chalfie shared the 2008 Nobel Prize in Chemistry for this work, and researchers now frequently use GFP and similar molecules as visual markers in experiments across species and cell types.
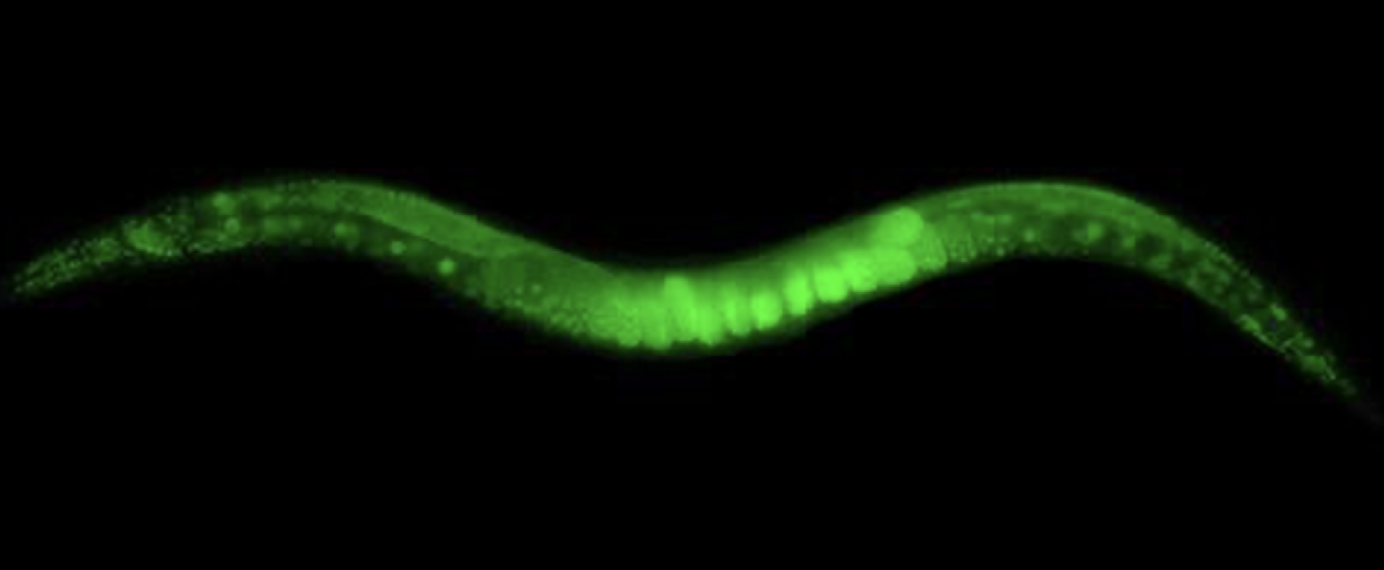
A C. elegans with GFP expression glows green.
“C. elegans” by Dan Dickinson, Goldstein lab, UNC Chapel Hill, CC BY-SA 3.0.
A model for aging
In 1993, Cynthia Kenyon at the University of California, San Francisco and colleagues discovered that mutations to a single gene, daf-2, along with the normal activity of a second gene, daf-16, could more than double the lifespan of C. elegans. Kenyon and others intrigued by this discovery would go on to use C. elegans to ask questions about the molecular mechanisms governing aging. Researchers have also studied how equivalent genes affect aging in other animals, including humans. [11]
A model for sex determination, reproduction, and development
C. elegans has been used to explore questions related to sex, reproduction, and development. Barbara Meyer, then at MIT, now at the University of California, Berkeley, discovered the mechanism of sex determination in the worm, and has uncovered mechanisms by which gene expression is regulated to compensate an animal having one or two X chromosomes. Other researchers have used C. elegans to make important discoveries about germ cells, the cells that give rise to eggs and sperm. Judith Kimble and John White, then at the MRC Laboratory of Molecular Biology, now at the University of Wisconsin–Madison discovered the first germline stem cell niche in C. elegans, which is the place where animals maintain a pool of stem cells with which to keep producing new germ cells over time. This finding had implications for fertility and regeneration research. Geraldine Seydoux at Johns Hopkins University has used C. elegans to investigate unique features of germ cells, as well as how sperm and egg interact and how the early embryo prepares to form a complex adult body.
C. elegans worms mating.
Courtesy of the Meyer lab.
Understanding sense of smell
Thanks in large part to C. elegans having such a well-mapped nervous system, the worm has been a common model for researchers studying how animals sense and respond to stimuli in their environments. Cori Bargmann, an alumna of Whitehead Institute, now at Rockefeller University, studies how C. elegans sense and process outside stimuli, how those stimuli can trigger changes in behavior, and how the brain can be rewired to modify behaviors over time. Bargmann’s research has particularly illuminated the worm’s sense of smell. She found the first evidence of a receptor for a specific smell, and her work more broadly shed light on how animals are able to recognize many different types of smells.
Researchers studied how worms move when searching for food, as seen here, and built a neuronal model that can predict their behavior.
Adapted from “Video 1: Forward-Pause-Forward transition” from William M Roberts, Steven B Augustine, Kristy J Lawton, Theodore H Lindsay, Tod R Thiele, Eduardo J Izquierdo, Serge Faumont, Rebecca A Lindsay, Matthew Cale Britton, Navin Pokala, Cornelia I Bargmann, Shawn R Lockery (2016), “A stochastic neuronal model predicts random search behaviors at multiple spatial scales in C. elegans,” eLife 5:e12572, https://doi.org/10.7554/eLife.12572.
A rich history of discovery
This is just a small sampling of the important discoveries that have been made in C. elegans. WormBook has compiled a list of many such achievements, including the discovery of multiple key molecules and pathways present across animals.
Worms at Whitehead Institute
Michael uses C. elegans to study microRNAs.

Michael working with the worms.
Madeleine Turner/ Whitehead Institute
What is a microRNA? MicroRNAs are tiny RNAs that regulate gene expression by pairing with messenger RNAs, the RNAs that function as intermediary scripts to turn genes into their protein products. This pairing causes the cell to break down the messenger RNA. The more quickly that a messenger RNA is destroyed, the more weakly its corresponding gene is expressed. MicroRNAs therefore play a very important role in managing the identity and function of cells.
C. elegans have long played an important role in microRNA research; in fact, microRNAs were first discovered in C. elegans. Victor Ambros and colleagues, and Gary Ruvkun and colleagues, published papers describing the first identified microRNA, lin-4, and its target, in 1993. At first, researchers thought that the small gene-regulating molecule might be an oddity. However, in 2000, Ruvkun discovered a second microRNA, and by the next year researchers—including Whitehead Institute Member David Bartel—had identified many more microRNAs in C. elegans, as well as microRNAs in other species. [12] Collectively, this research implied that microRNAs were a common and important regulator of gene expression across species. The field of microRNA research exploded, and microRNAs became the focus of Bartel’s lab.
In spite of the importance of C. elegans in establishing microRNA research, both in the field at large and in the Bartel lab specifically, no one in the lab was using C. elegans as a research model when Michael joined. However, as Michael–who had previous experience working with C. elegans–began to plan his graduate research, he realized that the worm would be the perfect model in which to explore his topic of interest: how microRNAs are regulated. Bartel agreed, and so C. elegans made their triumphant return to Whitehead Institute.
“It’s great to return to C. elegans,” says Bartel, who is also a professor of biology at MIT and a HHMI investigator. “Michael is working on sets of microRNAs that we discovered over 20 years ago. Since then, we and others have learned a lot about microRNAs, using a variety of research models. It will be fun to see what new things we will learn with C. elegans.”
Michael is using C. elegans to better understand how microRNAs, which degrade messenger RNAs, are themselves degraded. In recent years, researchers discovered a surprising mechanism of microRNA degradation: in some instances, when a microRNA pairs with a messenger RNA, instead of this leading to the destruction of the messenger RNA, it leads to the destruction of the microRNA.
“The normal regulatory logic is completely flipped,” Michael says. “This was discovered in mammalian cells, and our lab and others have been working out the mechanism for how this happens.”
The Bartel lab found that a particular gene is necessary for this process. When Michael joined the lab, he wondered whether that gene’s equivalent in C. elegans serves the same role. He found that it does. However, there are differences between how this process works in C. elegans and mammals, in particular in the way that the RNAs pair to trigger destruction of the microRNA. Those differences, and what they reveal about how microRNAs are regulated, are what Michael is studying now.
“What is the underlying principle of what's required for microRNAs to be degraded through this pathway in worms? It's not known, and that's what I'm trying to find out,” Michael says.
Michael hopes that what he learns from the worms will shed light on the logic for how microRNAs are regulated across animal species. This will in turn give researchers a better understanding of how cells are able to so precisely tailor their gene expression. The prospect of deciphering such a central facet of cell biology is exciting for Michael, but it’s just another day for C. elegans. Decades of results have shown that there’s no limit to what these simple worms can be used to discover.
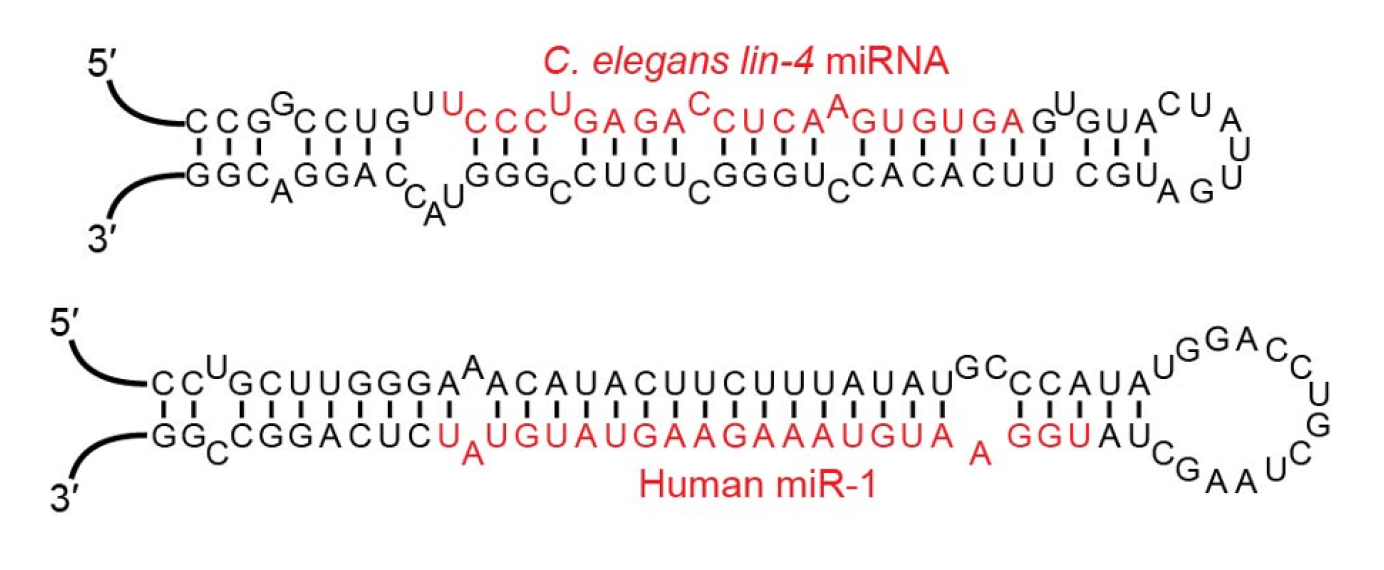
Two microRNAs in red, inside of their precursor structures. The top microRNA, C. elegans’ lin-4, was the first microRNA ever discovered.
“Examples of miRNA stem-loops, with the mature miRNAs shown in red” by VTD, CC BY-SA 4.0
Caenorhabditis elegans (C. elegans)
“Caenorhabditis elegans 02” by Heiti Paves, CC BY-SA 4.0.
[1] https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1213120/, Brenner Nobel speech
[2] Corsi, Ann K., Wightman, Bruce, Chalfie, Martin, A Transparent window into biology: A primer on Caenorhabditis elegans (June 18, 2015), WormBook, ed. The C. elegans Research Community, WormBook, doi: 10.1895/wormbook.1.177.1, http://www.wormbook.org
[3] Wormbook “Life cycle” chapter
[4] Wormbook “Sexual forms and their importance” chapter
[5] https://www.scientificamerican.com/article/c-elegans-connectome/
[6] https://www.nature.com/articles/s41586-019-1352-7
[7] https://www.sanger.ac.uk/news_item/1998-12-10-the-first-sequence-of-an-…
[8] https://www.science.org/doi/10.1126/science.282.5396.2011
[9] https://www.yourgenome.org/stories/the-pilot-project-for-the-human-geno…
[10] https://www.genome.gov/27552817/worm-genome-sequencing-influenced-human…
[11] https://www.pnas.org/doi/10.1073/pnas.1114658108
[12] https://www.sciencedirect.com/science/article/pii/S0092867404000455#BIB…
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu


