Scientists discover role for dueling RNAs
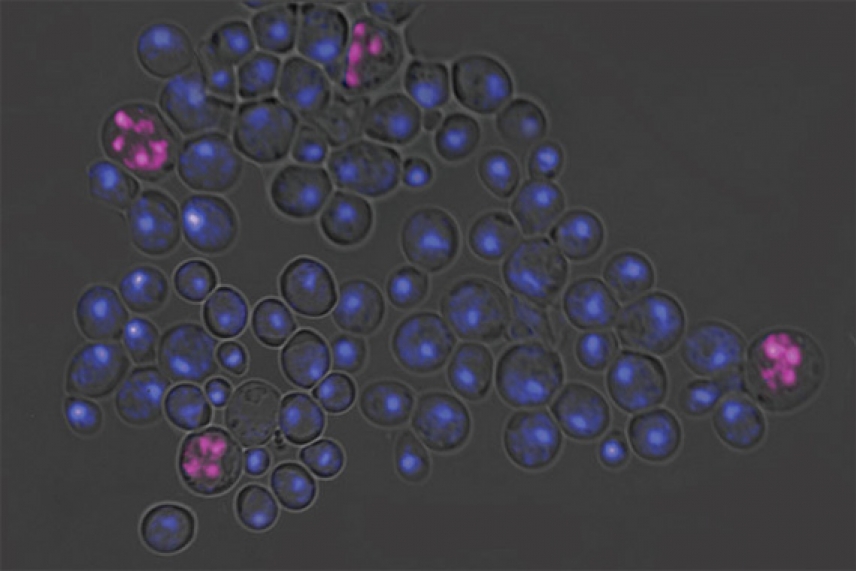
Why don't haploids make sense? Haploid cells, which contain one copy of each chromosome, should not undergo further reduction of their chromosome number (through a type of cell division called meiosis). To prevent such a catastrophic event, haploids block the production of a protein called IME4 by expressing antisense IME4. Failure to block sense IME4 results in the aberrant meiotic events shown left in pink.
Image by Cintia Hongay
CAMBRIDGE, Mass. — Researchers have found that a class of RNA molecules, previously thought to have no function, may in fact protect sex cells from self-destructing. These findings will be published in the November 17 issue of the journal Cell.
Central to this discovery is the fundamental process of gene expression. When a gene is ready to produce a protein, the two strands of DNA that comprise the gene unravel. The first strand produces a molecule called messenger RNA, which acts as the protein’s template. Biologists call this first strand of DNA the “sense” or “coding” transcript. Even though the other strand doesn’t contain a protein recipe, it may also, on occasion, produce an “anti-sense” RNA molecule, one whose sequence is complementary to that of the messenger, or sense, RNA. Antisense RNA has been detected for a number of genes, but is largely considered a genetic oddity.
Using common baker’s yeast, Cintia Hongay, a postdoctoral researcher in the lab of Whitehead Member and MIT Professor Gerald Fink, discovered that in the case of a gene called IME4, the antisense RNA blocks the sense RNA. In other words, the gene disables its own ability to make protein.
“This is the first case where a specific function in a higher cell for antisense RNA has been found,” says Fink, senior author on the paper. “This points to an entirely new process of gene regulation that we’ve never seen before in eukaryotic cells.”
There is a method to this sense/antisense madness, one that has a kind of yin and yang quality. When conditions around yeast cells are good and rich in nutrients, the cells divide by mitosis—that is, the DNA duplicates so each daughter cell receives exactly the same number of chromosomes as the original cell. However, when the yeast cells are starving, IME4 switches on and activates a process called meiosis. Here, the cells divide into germ-cell spores that, like mammalian egg and sperm cells, have half the number of chromosomes. Yeast spores withstand this harsh environment far more ably than the larger cells from which they originate.
But in some cases, flipping the meiotic switch can be catastrophic. If a cell with only one copy of each chromosome (a haploid cell) is forced into meiosis, the progeny won’t survive. Fortunately, such destructive meiotic division is avoided in haploid cells because they continually produce IME4 antisense RNA, blocking the production of sense RNA. Antisense IME4, then, safeguards against meiosis in cells that can’t handle it.
“This is the first time that we’ve found a function for antisense RNA, that is not RNAi, in a higher cell type,” says Hongay. “In fact, it’s really the first time we’ve seen a gene regulate itself in this way.”
“For years scientists have evaluated genomes by measuring the sense RNA, with antisense transcripts thought to have no meaning at all,” says Fink. “Here we’ve found a process in which antisense RNA regulates sense RNA. This same process may occur in the sex cells of mammals. In fact, considering how widespread these antisense transcripts are, I wouldn’t be surprised if these findings eventually lead us to discover an entirely new level of gene regulation.”
Hongay is now searching the yeast genome for other genes that might be regulated by antisense RNA.
This work was supported by the National Institutes of Health. Cintia Hongay is supported by a Ruth L. Kirschstein NRSA postodoctoral fellowship.
***
Hongay, C. F., Grisafi, P. L., Galitski, T., & Fink, G. R. (2006). Antisense Transcription Controls Cell Fate in Saccharomyces cerevisiae. Cell, 127(4), 735-745. doi:10.1016/j.cell.2006.09.038
Topics
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu


