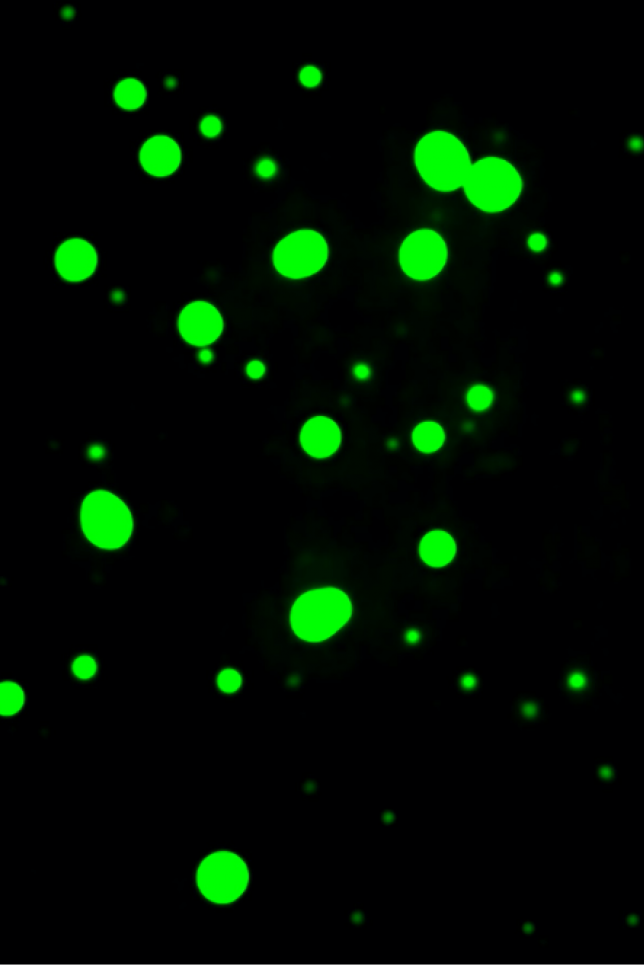
Researchers formed these droplets in the lab to investigate the role of RNA in their formation and dissolution.
Jon Henninger

MED1 condensates
Isaac Klein
RNAs: Assemble!
This story is part of our series, Sculptors of the Cell: RNA research at Whitehead Institute: RNA Research at Whitehead Institute. Click here to see all stories in this collection.
Some scientists hypothesize life on earth began with a single molecule: ribonucleic acid, or RNA. Made of chains of phosphate, sugar, and nucleic acids, RNA molecules could have formed and dissolved in the primordial soup until a self-replicating, ever-evolving RNA emerged, forming the basis upon which all of life was built.
This idea is called the “RNA World” hypothesis — and although it’s a debated topic, it’s undeniable that RNA molecules are some of the most dynamic and versatile molecules in living organisms today. They serve as the messengers that facilitate the creation of proteins from DNA sequences; they regulate how long other molecules can stay in cells; they can catalyze reactions between proteins, bringing together disparate actors to achieve a goal; and importantly, they can team up with other RNAs and proteins to help facilitate cellular processes.
DNA, RNA’s double-stranded cousin, is important mainly because of its sequence. RNA has a sequence too, but one reason these tiny molecules serve so many different functions in the cell is due to their physical properties. In fact, scientists know of thousands of RNAs that do not code for any proteins (many of their purposes are still unknown).
“RNAs can bind to themselves to make secondary structures, they can bind to other RNAs to make tertiary structures, there is so much variation in how they interact with each other — and much of this is not sequence specific,” says Whitehead Institute director Ruth Lehmann. “That’s why some people put RNAs at the center of the RNA World.”
In this article, we will venture beyond the sequence to explore some of the chemical and physical properties of RNA, and how those properties allow them to group with other RNAs to help the cell function — or cause disease.
Sticky tape molecules
To understand how RNA groups up in the cell, it helps to understand the basic properties of RNA molecules. Unlike DNA, which consists of two intertwined strands, RNA is a single stranded molecule. The backbone of the molecule is composed of alternating sugar and phosphate groups, and the other side — the “sticky side” — holds the nucleic acids adenine, guanine, cytosine and uracil like the teeth of a comb.
Imagine you’re holding a long piece of tape. If you bring your hands together, invisible forces pull that piece of tape in on itself, binding it together in twists and loops. That’s pretty much what happens with RNA strands in the cell, says Whitehead Institute Member Ankur Jain. “Nucleic acids can form base pairs, A can base pair with U, G can base pair with C,” Jain says. “There are many, many sites where a single RNA molecule can bind to itself.”
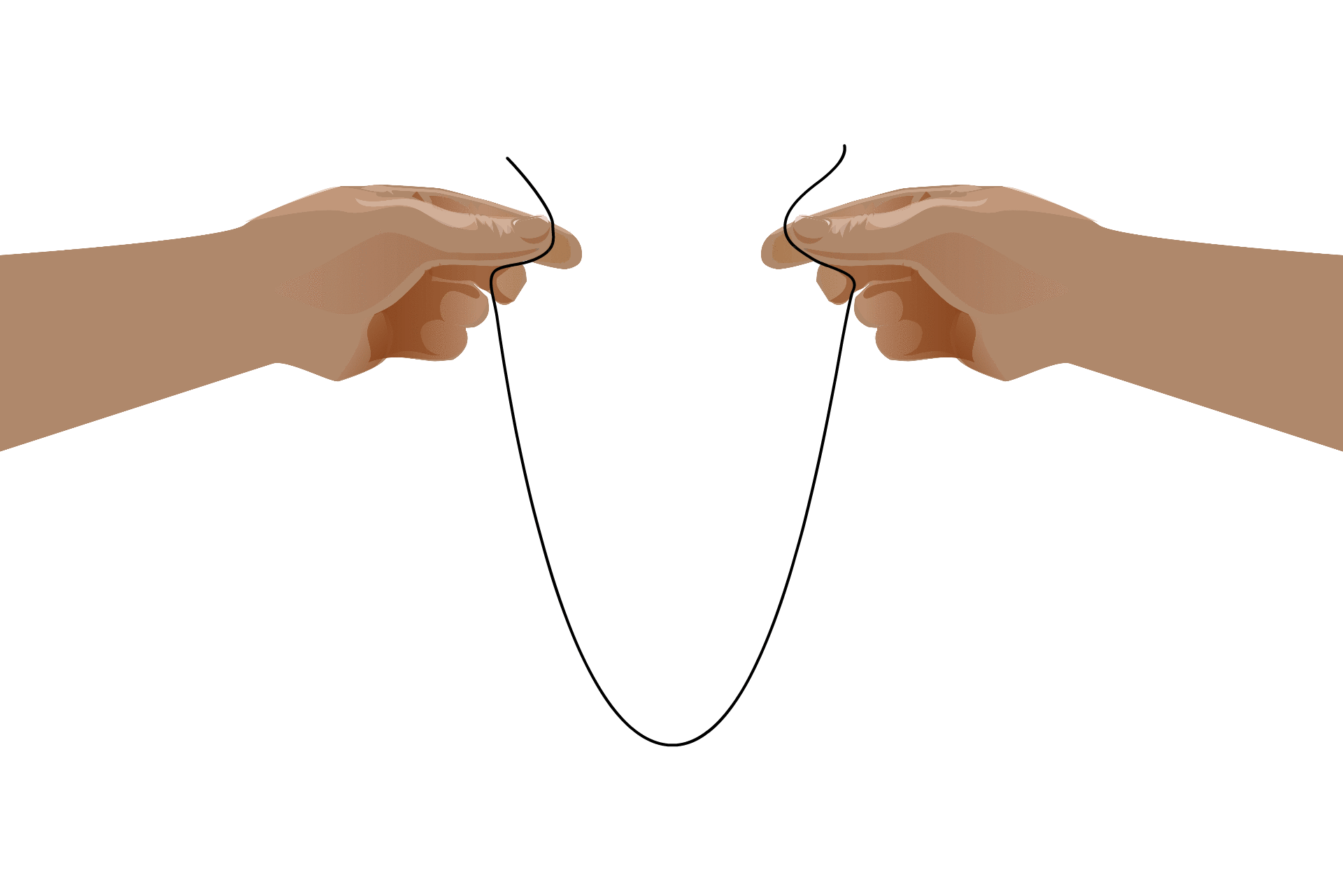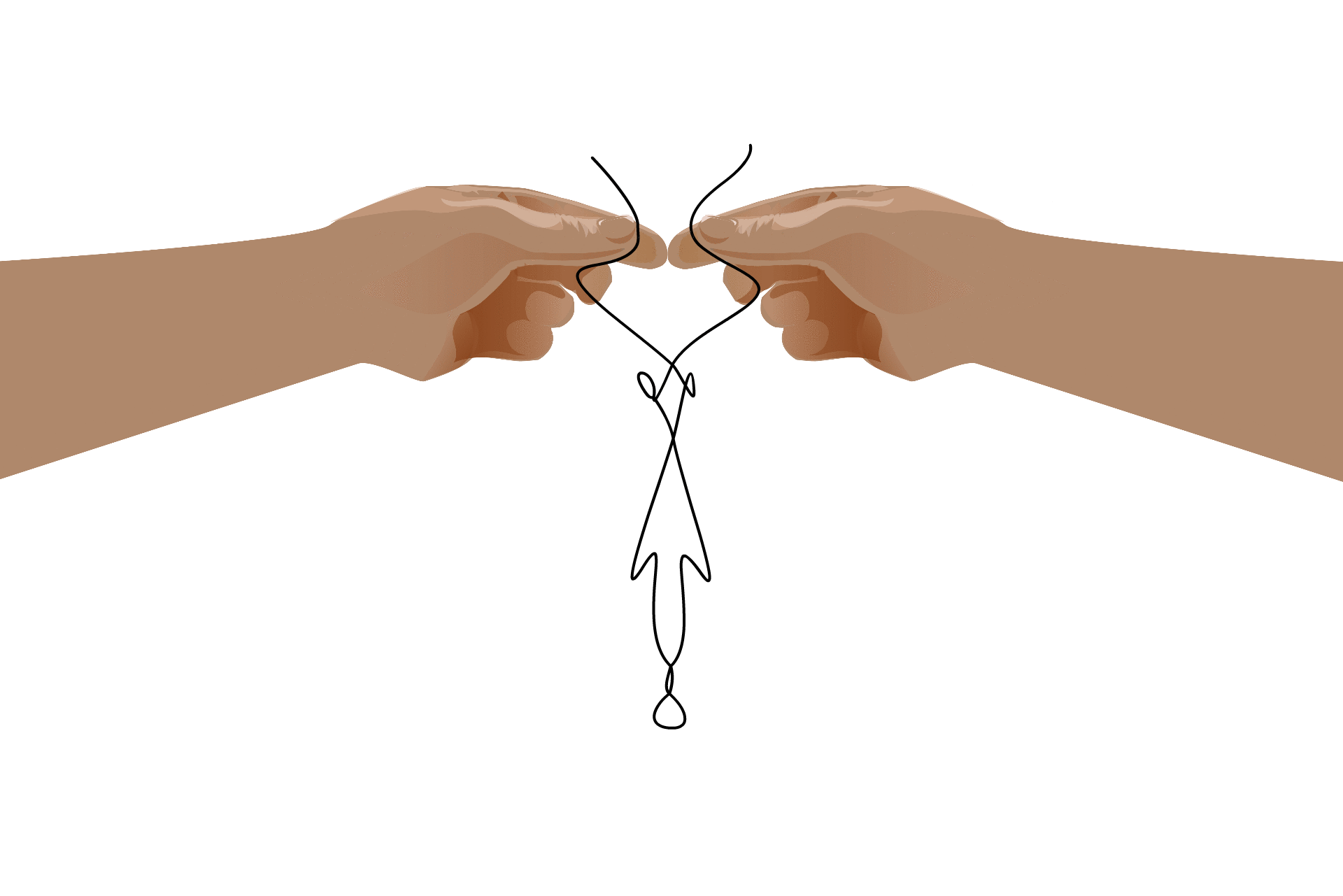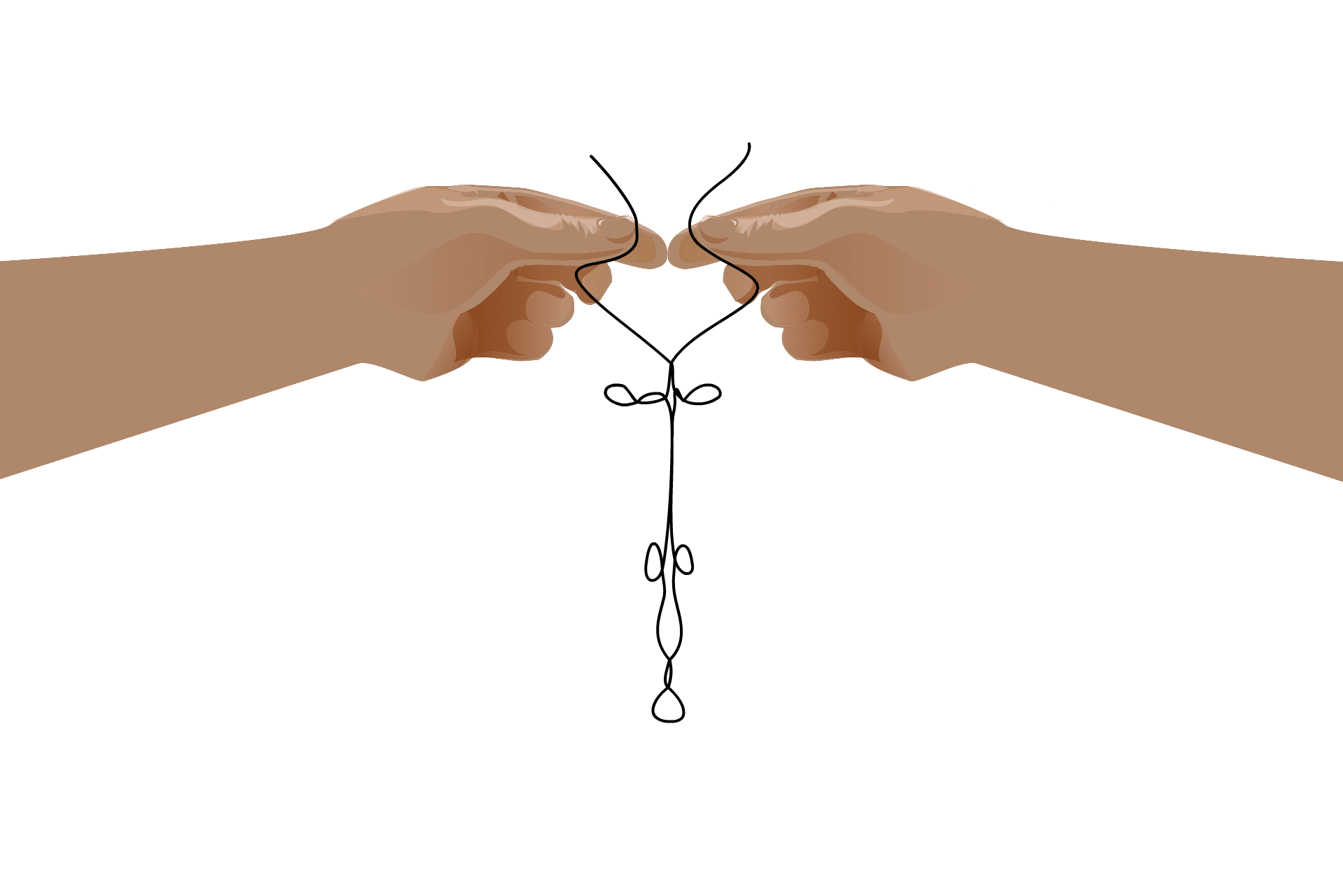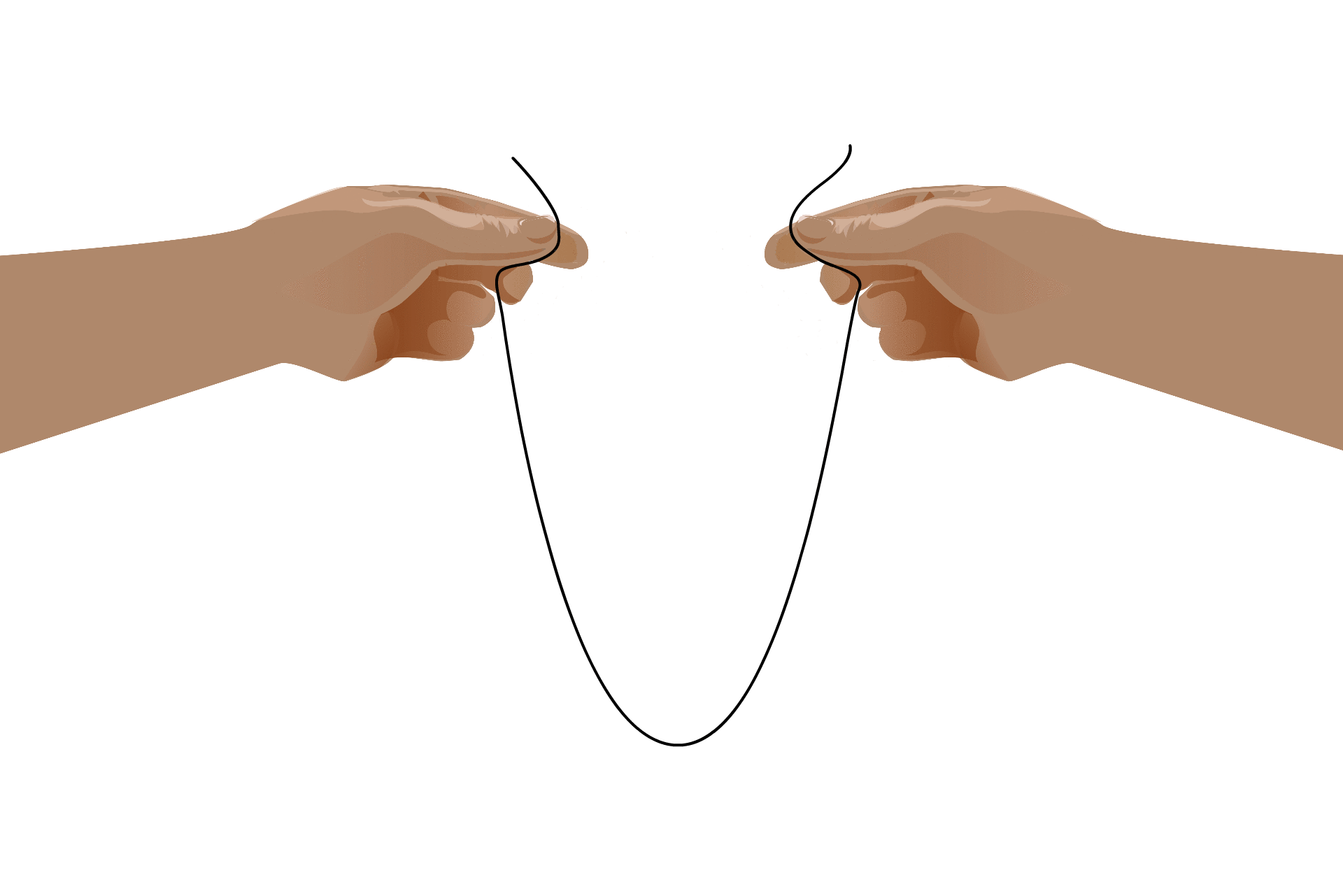
This self-binding leads to an RNA’s “secondary structure,” which is necessary for some processes that take place in the cell. Although the formation of these structures is due to the RNA’s nucleotide sequence, once the structure has formed they take on a life of their own. Some RNAs, called ribozymes, fold into useful shapes that assist with chemical reactions in the cell.
When good RNA goes bad
The same properties that make RNA perfect for creating useful secondary structures can sometimes play a role in diseases — especially when mutations in the DNA lead to numerous repetitive sequences. Ankur Jain studies what happens when this type of mutation, called a repeat expansion, occurs in the human body.
Repeat expansions can happen via DNA replication errors on various short repeats that litter the human genome. This form of mutation makes the affected section of DNA much longer than before. That, in turn, leads to very long RNA molecules when the DNA is transcribed.
These super-long RNAs are sticky, just like regular RNAs — except with all that extra bulk it’s easy for them to get tangled up together. “It's the same physical principle by which hair or a long floppy piece of rope gets tangled, here applied at a molecular level,” says Jain.
These tangles can compound on each other and form harmful, irreversible aggregates. “These aggregates of RNA can clump up cellular proteins,” Jain says. “They act like pieces of sponge, and sequester other proteins inside the cell.”
This can contribute all sorts of problems in cellular functions — and when RNA aggregates occur in cells in the nervous system, they may contribute to diseases such as ALS, Huntington disease and Fragile X syndrome. Jain and his lab are studying how these aggregates might play a role in diseases, and are searching for ways to disrupt or dissolve them in cells.
RNAs of a feather…
While these irreversible aggregates of RNA can be harmful to cells, there is another type of grouping involving RNAs that is actually very necessary for the cell to complete functions such as transcription. “The flip side of RNA self assembly is these non-membrane bound bodies inside the cell that contain RNA and RNA binding proteins,” Jain says.
These groupings, called condensates, are small, membraneless organelles that can form and dissolve quickly and easily, unlike the harmful aggregates that Jain studies. Condensates are often composed of RNAs, proteins called RNA binding proteins, and other cellular components that differ depending on the function of the condensate. Scientists have likened these condensates to droplets of oil in a glass of water.
Over the past decades, new information on condensates has shaped the way we understand cell biology. Many students learned about the inside of a cell as a kind of soup, with all the necessary components for cellular functions mixed together and spread evenly throughout the cytoplasm. Now, scientists recognize that the cell is constantly creating and dissolving condensates in order to accomplish specific functions like transcription, RNA splicing or the creation of ribosomes.
There are several labs at Whitehead Institute that investigate different kinds of condensates. In 2018, Whitehead Institute Member Richard Young and his lab discovered a new kind of droplet called a transcriptional condensate. These new condensates bring together all the different proteins needed to successfully transcribe a gene.
Two years later, in 2020, the Young lab discovered something interesting about the effect of RNA on these condensates: RNA molecules themselves — the products of transcription — are responsible for regulating their formation through a feedback loop. Too few RNA molecules, and the cell initiates transcription to create more. Too many, and transcription draws to a halt.
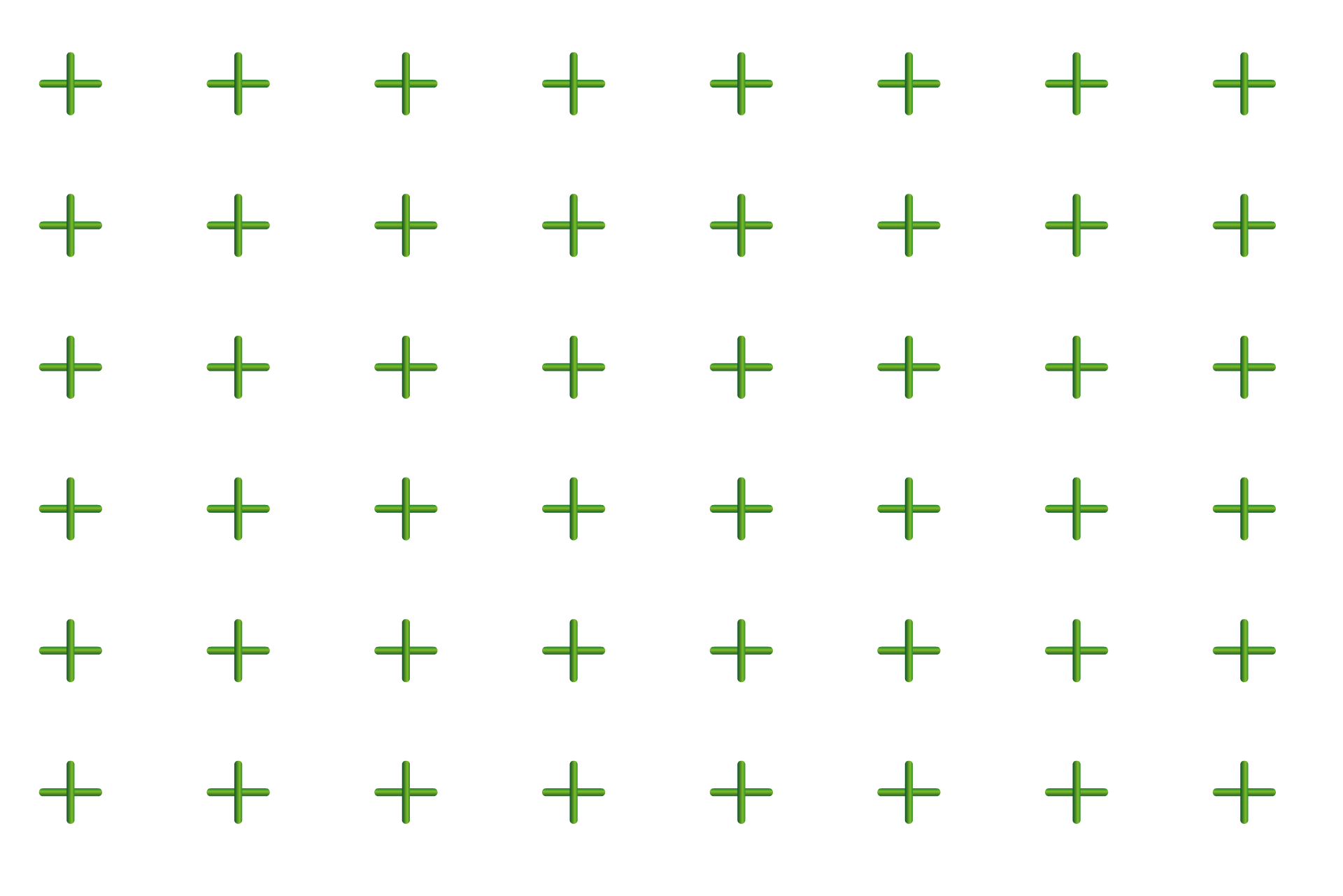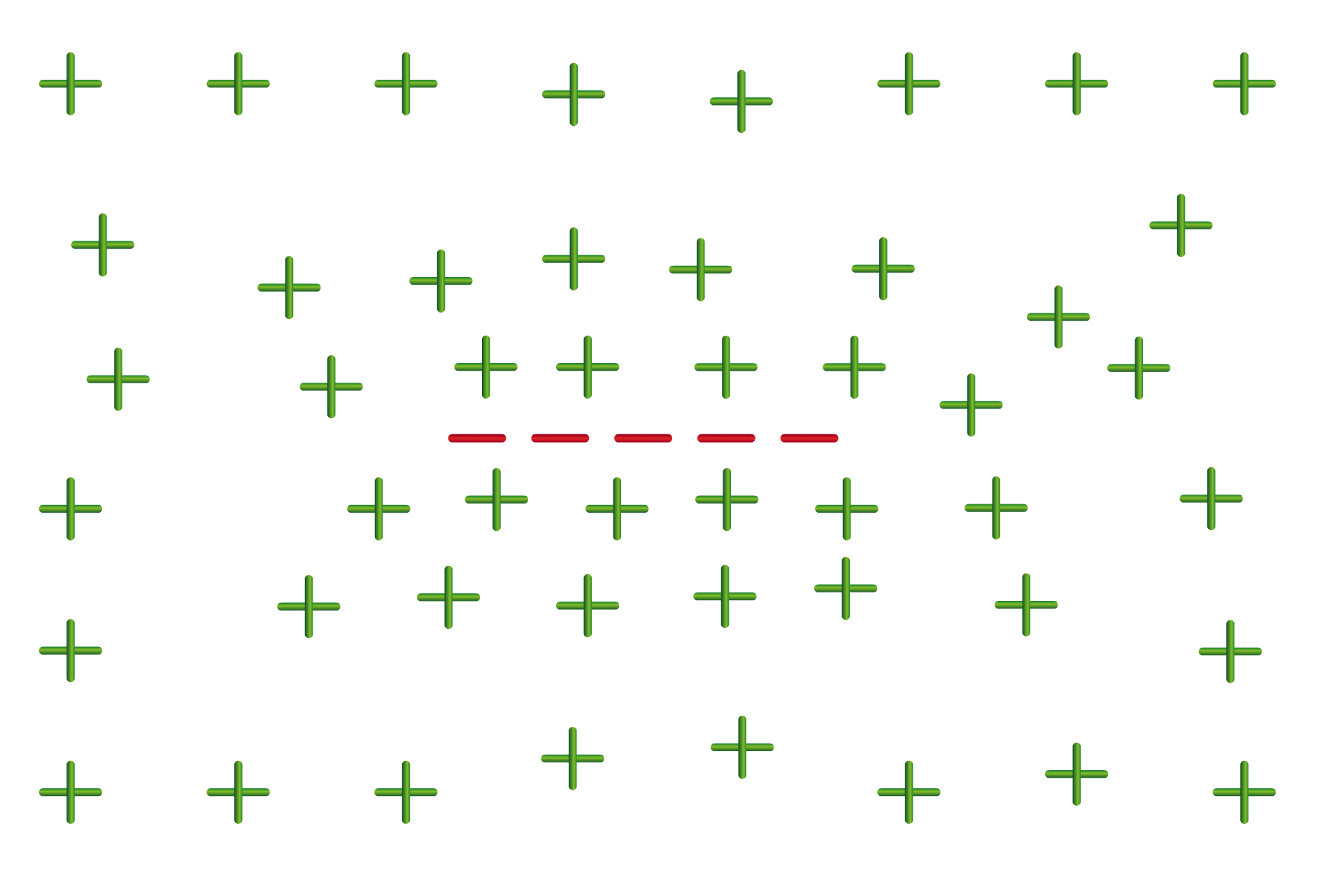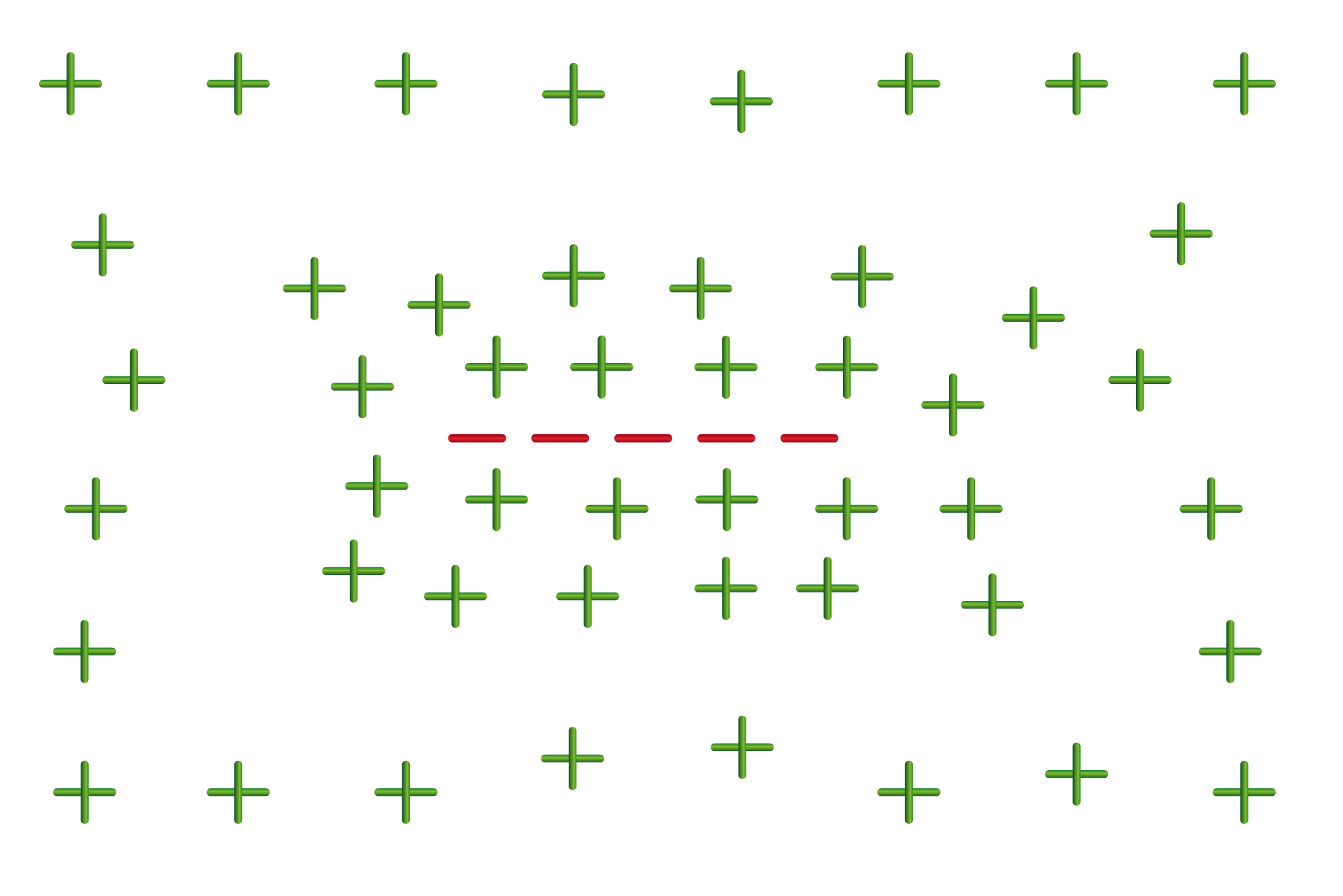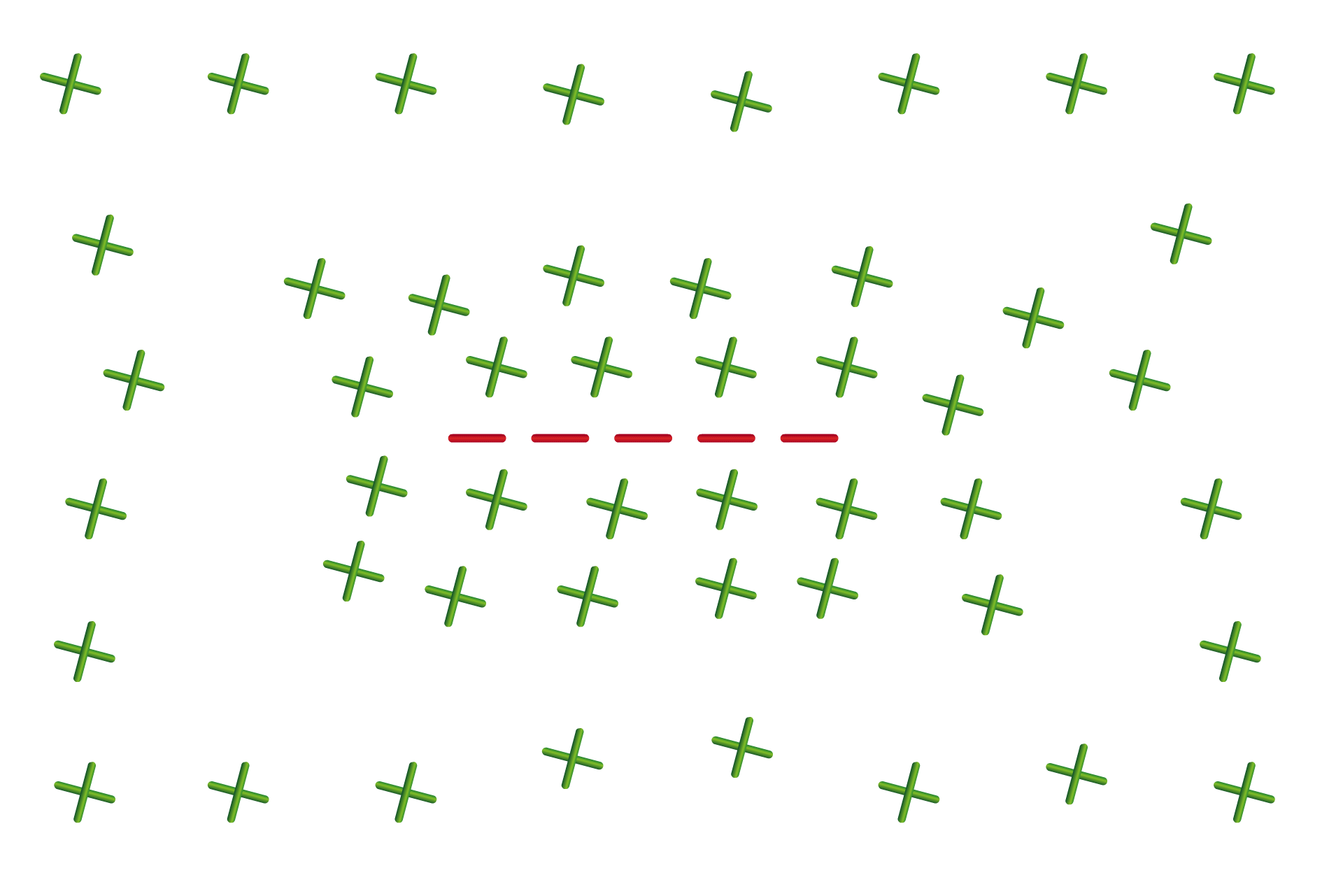
This behavior was independent of the sequence of the RNA molecules, and instead depended on another of their properties: their charge. While one side of an RNA molecule holds the nucleotides, the other side is composed of a “backbone” of alternating sugar and phosphate molecules. Many RNA interactions are due to this part of the molecule, which carries an extremely negative charge.
“Many proteins, at the pH that you have in biology, will have a net positive charge,” Young says. “And because of this, they tend to repel one another. But if you have another polymer like RNA that is just a string of negative charges, it can neutralize that repulsion, and help things come together. Where the RNA balances that net positive charge of the proteins is where you get the maximum amount of condensate formation.”
Young likens this to the way lightning forms in a storm. The bottom of the storm cloud is full of negative charges, and the ground below is more positive — and in order to balance that charge, many negative charges join together in a crackling bolt to meet positive charges from the ground.
Thinking of RNA in terms of charge instead of sequence has led Young and his lab to wonder whether some of RNAs encoded in the genome that don’t code for proteins might instead have a function in controlling condensates.
“Our big question right now is, what is the function of the long noncoding RNAs in cells?” Young says. “Researchers have thought that many don't have a function, but what we're imagining now is that these RNAs tune the transcriptional condensates at the places on the genome where they’re being transcribed. We're just starting work on that.”
Partner proteins
While Young’s lab focuses on transcriptional condensates, Whitehead Institute director Ruth Lehmann studies a different type of condensate, called a germ granule. Made from various RNA and protein molecules, these granules are believed to regulate the translation of messenger RNA (mRNA) molecules inside germ cells — the cells that give rise to eggs and sperm — during development.
Lehmann first became interested in germ granules during her graduate work on genes that affected the patterning of embryos. She had noticed a few proteins in germ cells that were essential for proper formation of the embryo— and one in particular that seemed to be important in creating the one of the two “poles” of the egg (these poles set the front to back spatial blueprint for the embryo to develop later on).
After conducting a few experiments, she realized that this protein, which she named Oskar, was an RNA binding protein that served as the life of the germ granule party; if Lehmann altered the Oskar protein so it collected at the opposite side of the germ cell, all the other “party guests” — RNAs and proteins essential for germ cell function — followed.
As it turned out, Oskar is serving as the “seed” protein for the germ granules by binding a few specific RNAs and other RNA binding proteins , which in turn bind more proteins and attract other RNAs. This mechanism of condensate formation shows a different method of condensate formation — one in which RNA charge still plays a role, but is aided by proteins like Oskar.
“What's beautiful about Ruth’s germ cell system is that the proteins that she's been studying help set up one pole of the developing embryo — and Oskar has a RNA binding domain that sequesters a variety of RNAs at that site and it forms a condensate through that mechanism,” Young says. “So there are at least two mechanisms: one involves specific RNA binding proteins, and one involves the charge balanced properties of proteins and RNA.”
Transcriptional condensates and germ granules are not the only condensates that can form in cells. Whitehead Institute Member David Bartel, for example, studies another kind of condensate called P granules. Researchers are still discovering new roles for RNA in the formation of condensates.
Whitehead Institute’s own “RNA World”
Over the past few years, Whitehead Institute has become somewhat of an RNA hotspot. From Whitehead Fellow Silvi Rouskin’s work on secondary structure, to David Bartel’s focus on small RNAs called microRNAs, researchers at the Institute are tackling questions about RNA and condensate biology from a number of angles.
“There are so many synergies that we can draw upon at Whitehead Institute,” says Ankur Jain. “For instance, Silvi's lab is studying RNA structure, and if I was at a different place, it would have been very difficult for me to adopt her technologies to study RNA aggregates. But now students in my lab can just walk over to her lab, and learn the tricks that she has been using to study its structure in vivo. It's really stimulating to be at the forefront of RNA research.”
Topics
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu


