Prions play powerful role in the survival and evolution of wild yeast strains
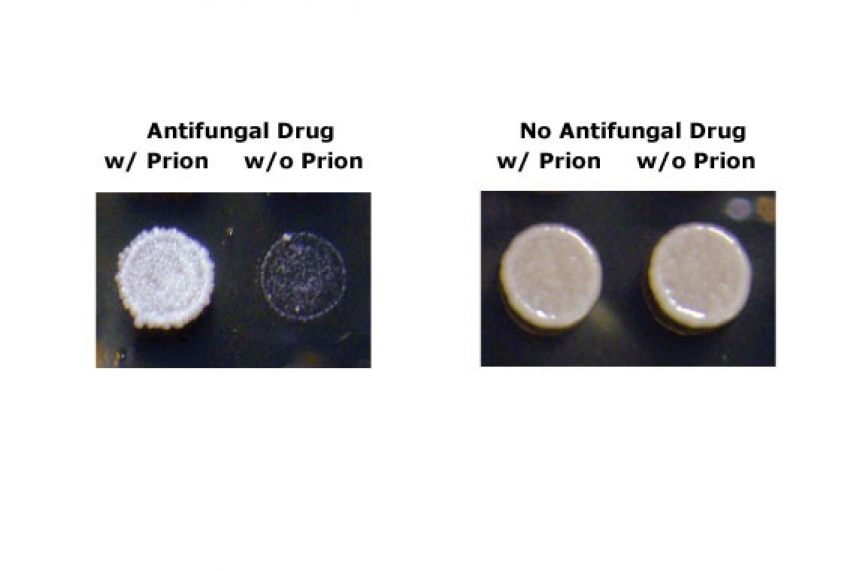
Yeast with prions grow better than those without when challenged with an antifungal drug.
Courtesy of Nature, R. Halfmann, D. F. Jarosz, S. K. Jones, A. Chang, A. K. Lancaster, and S. Lindquist
CAMBRIDGE, Mass. – Prions, the much-maligned proteins most commonly known for causing "mad cow" disease, are commonly used in yeast to produce beneficial traits in the wild. Moreover, such traits can be passed on to subsequent generations and eventually become "hard-wired" into the genome, contributing to evolutionary change.
Prions were first found to produce heritable new traits more than a decade ago in laboratory studies of simple baker's yeast. The key discovery then was that some proteins could spontaneously switch from a normal shape into a self-perpetuating prion conformation. The switch to the prion state alters protein function, which can result in the appearance of new traits, some helpful, some detrimental. Sophisticated cellular machinery ensures that replicating prion templates are chopped into pieces that can be passed to daughter cells during cell division. Importantly, the rate at which proteins switch into and out of the prion state increases in response to environmental stress, suggesting that they are part of an inherent survival mechanism that helps yeasts adapt to changes in their surroundings.
Yet, as compelling as the case for this protein-based mechanism of inheritance is, its biological significance has been hotly debated for one key reason: prions capable of modifying phenotypes have never been found in nature. Until now.
In a massive undertaking, Whitehead Institute scientists have tested nearly 700 wild yeast strains isolated from diverse environments for the presence of known and unknown prion elements, finding them in one third of all strains. All the prions appear capable of creating diverse new traits, nearly half of which are beneficial. These unexpected findings, reported in this week's edition of the journal Nature, stand as strong evidence against the common argument that prions are merely yeast "diseases" or rare artifacts of laboratory culture.
"A huge amount of effort has gone into studying this paradigm-shifting mode of inheritance, but with no real understanding of whether it's genuinely important biologically," says Daniel Jarosz, co-first author of the Nature paper and a postdoctoral researcher in the lab of Whitehead Member Susan Lindquist. "Now it seems clear they do influence the way natural yeasts cope with changing environments and evolve in response to stress."
The hunt for prions in wild yeast strains began in the Lindquist lab when Jarosz and Randal Halfmann, then an MIT graduate student and now a fellow at the University of Texas Southwestern Medical Center, gathered hundreds of wild strains from stock centers all over the world. They then conducted a chemical screen for one well-studied prion, [PSI+], and found it in 10 wild strains. Genetic manipulations confirmed its status as a true prion. They then observed the effects of [PSI+] on biological traits by eliminating the prion conformation in these strains and exposing them to natural stresses, such as high acidity and the presence of antifungal agents. In one strain isolated from Beaujolais wine, for example, the prion resulted in the emergence of traits that could be beneficial or detrimental, depending upon environmental conditions. Another well-known prion, [MOT3+] was found in six wild strains.
To determine whether the wild yeasts might harbor other unknown prion elements, Jarosz and Halfmann exposed sets of cultures of all the wild strains to the same chemical protocol that switched [PSI+] and [MOT3+] out of their prion states. In all, 255 strains demonstrated different phenotypes under varying stressors after this treatment.
"We certainly didn't expect to see this much prion-based phenotypic diversity," Jarosz says. "It's remarkable."
Another surprise was that approximately 40% of the traits produced by the wild prions proved to beneficial to growth in the dozen different environmental conditions tested.
"How frequently beneficial they are suggests that the prions have already been subject to previous, positive selective events," says Lindquist. "We see them as part of a bet-hedging strategy that allows the yeast to alter their biological properties quickly when their environments turn unfavorable."
Convinced of the impact prions have had on yeast evolution, Lindquist speculates that these shape-shifting proteins may be "remnants of early life," from a time when inheritance was predominantly protein-based rather than nucleic-acid based. She also theorizes that prions may play such roles beyond yeast, and her lab intends to take similar approaches in the hunt for prion activity in other organisms.
This work was supported by grants from the G. Harold and Leila Y. Mathers Foundation, HHMI, the Damon Runyon Cancer Research Foundation and the National Institutes of Health (NIH Pathway to independence award).
* * *
Susan Lindquist's primary affiliation is with Whitehead Institute for Biomedical Research, where her laboratory is located and all her research is conducted. She is also a Howard Hughes Medical Institute investigator and a professor of biology at Massachusetts Institute of Technology.
* * *
Citation:
Halfmann, R., Jarosz, D. F., Jones, S. K., Chang, A., Lancaster, A. K., & Lindquist, S. (2012). Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature, 482(7385), 363-368.
Whitehead Institute for Biomedical Research is a nonprofit, independent research and educational institution. Wholly independent in its governance, finances and research programs, Whitehead shares a close affiliation with Massachusetts Institute of Technology through its faculty, who hold joint MIT appointments.
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu


