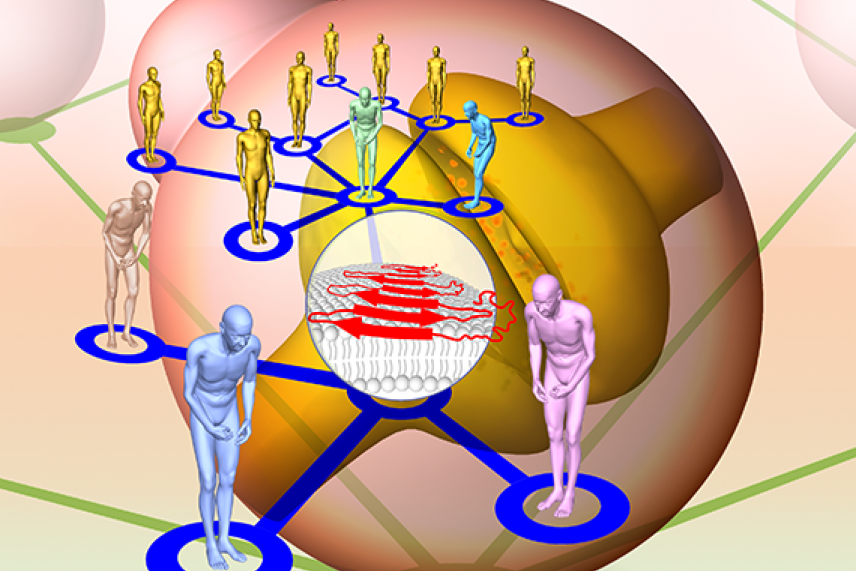
The authors built molecular networks in yeast cells and in neurons to draw relationships between different genetic forms of Parkinson's (stylized as patients in different colors) and the aggregation of the protein alpha-synuclein (at center in red).
Steven Moskowitz
New Clues on the Basis of Parkinson’s Disease and Other “Synucleinopathies”
Cambridge, Mass., —Parkinson’s disease (PD) and other “synucleinopathies” are known to be linked to the misfolding of alpha-synuclein protein in neurons. Less clear is how this misfolding relates to the growing number of genes implicated in PD through analysis of human genetics. In two studies published in the advance online edition of Cell Systems, researchers affiliated with Whitehead Institute and Massachusetts Institute of Technology (MIT) explain how they used a suite of novel biological and computational methods to shed light on the question.
To start, they created two ways to systematically map the footprint of alpha-synuclein within living cells. “In the first paper, we used powerful and unbiased genetic tools in the simple Baker’s yeast cell to identify 332 genes that impact the toxicity of alpha-synuclein,” explained Vikram Khurana, first and co-corresponding author on the studies. “Among them were multiple genes known to predispose individuals to Parkinson’s—so we show that various genetic forms of Parkinson’s are directly related to alpha-synuclein. Moreover, the results showed that many effects of alpha-synuclein have been conserved across a billion years of evolution from yeast to human,” said Khurana, former Visiting Scientist at the Whitehead Institute.
“In the second paper, we created a spatial map of alpha-synuclein, cataloging all the proteins in living neurons that were in close proximity to the protein,” explained Chee Yeun Chung, former Whitehead Institute Senior Research Scientist, who co-led both studies with Khurana. The mapping was achieved without disturbing the native environment of the neuron, by tagging alpha-synuclein with an enzyme—APEX—that allowed proteins less than 10 nanometers away from synuclein to be marked with a trackable fingerprint. “As a result, for the first time, we were able to visualize the protein’s location, at minute scale, under physiologic conditions in an intact brain cell,” noted Chung, who is now Scientific Co-founder and Associate Director at Yumanity Therapeutics in Cambridge.
Remarkably, the maps derived from these two processes were closely related and converged on the same Parkinson’s genes and cellular processes. Whether in a yeast cell or in a neuron, alpha synuclein directly interfered with the rate of production of proteins in the cell, and the transport of proteins between cellular compartments. “It turns out the mechanisms of toxicity of the misfolded protein are closely related to which proteins it directly interacts with, and that these interactions can explain connections between different Parkinson’s genetic risk factors,” said Khurana, now a Principal Investigator within the Ann Romney Center for Neurologic Diseases at Brigham and Women’s Hospital and the Harvard Stem Cell Institute.
Finally, the authors addressed two major challenges for any study that generates large data-sets of individual genes and proteins in model organisms like yeast: How to assemble the data into coherent maps? And how to integrate information across species, in this case from yeast to human?
Enter computational biologist Jian Peng, former Visiting Scientist at Whitehead Institute and postdoctoral researcher at MIT. “First, we had to figure out much better methods to find human counterparts of yeast genes, and then we had to arrange the humanized set of genes in a meaningful way,” explained Peng, now Assistant Professor of Computer Sciences at University of Illinois, Urbana-Champaign. “The result was TransposeNet, a new suite of computational tools that uses machine learning algorithms to visualize patterns and interaction networks based on genes that are highly conserved from yeast to humans—and then makes predictions about the additional genes that are part of the alpha-synuclein toxicity response in humans.”
This analysis produced networks that mapped out how alpha-synuclein is related to other Parkinson’s genes through well-defined molecular pathways. “We now have a system to look at how seemingly unrelated genes come together to cause Parkinson’s and how they are related to the protein that misfolds in this disease,” said Khurana. To confirm their work, the researchers generated neurons from Parkinson’s patients with different genetic forms of the disease. They showed that the molecular maps generated from their analyses allowed them to identify abnormalities shared among these distinct forms of Parkinson’s. Prior to this, there was no obvious molecular connection between the genes implicated in these varieties of PD. "We believe these methods could pave the way for developing patient-specific treatments in the future,” Khurana observed.
Research was supported by an HHMI Collaborative Innovation Award (V.K., C.Y.C., A.Y.T., and S.L.), the JPB Foundation (V.K., C.Y.C., S.L.), NIH K01AG038546 (C.Y.C.), U01CA184898 and R01GM089903 (E.F.), R01GM081871 (B.B.) and HG001715 (M.V. and D.E.H), an American Brain Foundation and Parkinson’s Disease Foundation Clinician-Scientist Development Award (V.K.), the Harvard Neurodiscovery Center Pilot Project Program (V.K.), the Multiple System Atrophy Coalition (V.K.), the Eleanor Schwartz Charitable Foundation (S.L.) and the NIH/NHGRI NHGRIP50HG004233 (M.V).
Potential Conflicts of Interest: V.K., C.Y.C., and S.L. are scientific co-founders of Yumanity Therapeutics, a company focused on developing neurodegenerative disease therapeutics.
The work of Chung, Khurana, and Peng discussed in this release was primarily conducted in the lab of Whitehead Institute Member, MIT Professor of Biology and HHMI Investigator, Dr. Susan Lindquist, who died Oct. 27, 2016.
* * *
Full Citation:
1] In situ proteomic approaches connect alpha synuclein directly to endocytic trafficking and mRNA metabolism in neurons
2] Genome-scale networks link neurodegenerative disease genes to alpha-synuclein through specific molecular pathways
Cell Systems, online January 25, 2017
“In situ proteomic” authors:
Authors: Chee Yeun Chung1†X*, Vikram Khurana1,2†*, Song Yi3§, Nidhi Sahni3¶, Ken H. Loh4, Pavan K. Auluck1,#, Valeriya Baru1, Namrata D. Udeshi5, Yelena Freyzon1, Steven A. Carr5, David E. Hill3, Marc Vidal3, Alice Y. Ting4‡, and Susan Lindquist1 6,7
Affiliations:
1Whitehead Institute for Biomedical Research, Cambridge, MA 02142, USA.
2Ann Romney Center for Neurologic Disease, Department of Neurology, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA 02115, USA, and Harvard Stem Cell Institute, Cambridge, MA 02138, USA.
3Center for Cancer Systems Biology (CCSB) and Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA 02215, and Department of Genetics, Harvard Medical School, Boston, MA 02115, USA.
4Department of Chemistry, Massachusetts Institute of Technology, Cambridge, MA 02142, USA.
5The Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
6Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
7Howard Hughes Medical Institute, Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
†These authors contributed equally to this work (co-first and co-corresponding authors).
X Current address: Yumanity Therapeutics, Cambridge MA 02139
¶ Current address: Department of Systems Biology, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA .and Graduate Program in Structural and Computational Biology and Molecular Biophysics, Baylor College of Medicine, Houston, TX 77030, USA.
§ Current address: Department of Systems Biology, The University of Texas MD Anderson Cancer center, Houston, TX 77030
# Current address: Biogen, Inc. Cambridge, MA 02142
‡ Current address: Dept. of Genetics, Biology and Chemistry, Stanford University, Stanford, CA 94305
*Correspondence to Chee Yeun Chung (cychung@yumanity.com; Lead Contact) and Vikram Khurana (vkhurana@bwh.harvard.edu)
“Genome-scale” authors:
Authors: Vikram Khurana*1,2, ‡, Jian Peng*1,3, X, Chee Yeun Chung*1,Y, Pavan K. Auluck1, Saranna Fanning1, Daniel F. Tardiff1,Y, Theresa Bartels1, Martina Koeva1,4, Stephen W. Eichhorn1 , Hadar Benyamini, Yali Lou1, Andy Nutter-Upham1, Valeriya Baru1, Yelena Freyzon1, Nurcan Tuncbag4, Michael Costanzo5, Bryan-Joseph San Luis5, David C. Schöndorf6, M. Inmaculada Barrasa1, Sepehr Ehsani1, Neville Sanjana,7,8, Quan Zhong9, Thomas Gasser6, David P. Bartel1, Marc Vidal10, Michela Deleidi6, Charles Boone5, Ernest Fraenkel4,‡, Bonnie Berger3,‡, Susan Lindquist1,11.
Affiliations:
1Whitehead Institute for Biomedical Research, Cambridge, MA 02142. USA.
2Ann Romney Center for Neurologic Disease, Department of Neurology, Brigham and Women’s Hospital and Harvard Medical School, Boston, MA 02115, USA, and Harvard Stem Cell Institute, Cambridge, MA 02138, USA.
3Computer Science and Artificial Intelligence Laboratory and Department of Mathematics, MIT, Cambridge, MA 02139, USA.
4Department of Biological Engineering, MIT, Cambridge, MA 02139, USA.
5Banting and Best Department of Medical Research, University of Toronto, Toronto, ON M5G 1L6, Canada.
6Department of Neurodegenerative Diseases, German Center for Neurodegenerative Diseases (DZNE), and Hertie-Institute for Clinical Brain Research, University of Tübingen, Tübingen, 72076, Germany.
7Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
8New York Genome Center and Department of Biology, New York University New York, NY 10013
9Department of Biological Sciences, Wright State University, Dayton, OH 45435, USA. 10Center for Cancer Systems Biology (CCSB) and Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA 02215, and Department of Genetics, Harvard Medical School, Boston, MA 02115, USA.
11HHMI, Department of Biology, MIT, Cambridge, MA 02139. USA.
X Current Address: University of Illinois, Department of Computer Science, University of Illinois at Urbana-Champaign, IL 61801, USA.
Y Current Address: Yumanity Therapeutics, Cambridge, MA 02139, USA.
*Equal contribution (co-first authors) ‡Correspondence: Dr Vikram Khurana vkhurana@bwh.harvard.edu (Lead Contact); Dr Bonnie Berger bab@csail.mit.edu; Dr Ernest Fraenkel fraenkel@mit.edu
Topics
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu
