Enhanced-sensitivity NMR could reveal clues on how proteins fold
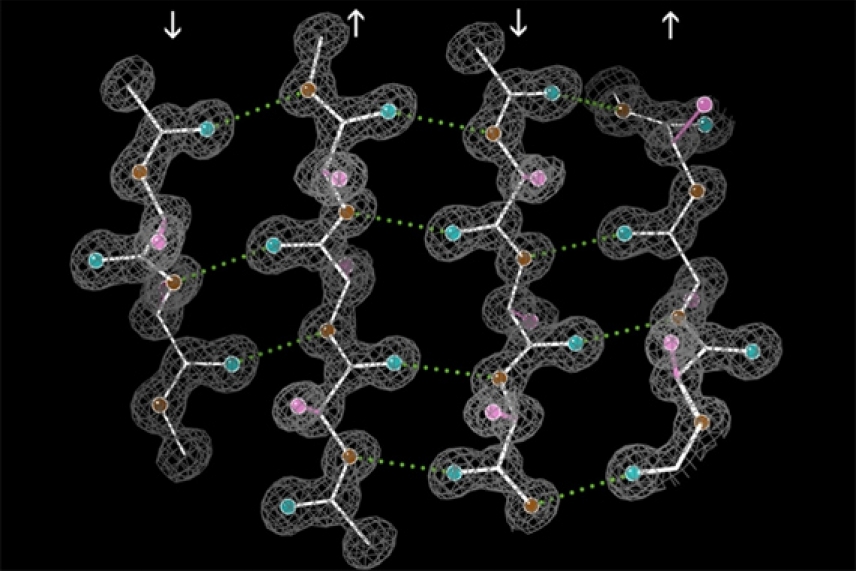
Antiparallel beta-sheet structure of the enzyme catalase: The antiparallel hydrogen bonds (dotted) are between peptide NH and CO groups on adjacent strands. Arrows indicate the chain direction, and electron density contours outline the non-H atoms.
Wikimedia Commons (edited by MIT News)
Proteins can fold in different ways depending on their environment. These different configurations change the function of the protein; misfolding is frequently associated with diseases such as Alzheimer’s and Parkinson’s.
Until now, it has been difficult to fully characterize the different structures that proteins can take on in their natural environments. However, using a new technique known as sensitivity-enhanced nuclear magnetic resonance (NMR), Whitehead Institute and MIT researchers have shown that they can analyze the structure that a yeast protein forms as it interacts with other proteins in a cell.
Using this type of NMR, which is based on a technique known as dynamic nuclear polarization (DNP), scientists can gain much more insight into protein structure and function than they can with current NMR technology, which requires large quantities of purified proteins, isolated from their usual environment.
“Dynamic nuclear polarization has a capacity to transform our understanding of biological structures in their native contexts,” says Whitehead Institute Member Susan Lindquist, who is also a professor of biology at MIT and one of the senior authors of the paper, which appears in the October 8 issue of Cell.
Robert Griffin, an MIT professor of chemistry and director of the Francis Bitter Magnet Laboratory, is also a senior author of the paper. Kendra Frederick, a former Whitehead postdoc who is now an assistant professor at the University of Texas Southwestern, is the paper’s lead author.
Heightened sensitivity
Traditional NMR uses the magnetic properties of atomic nuclei to reveal the structures of the molecules containing those nuclei. By creating a magnetic field that interacts with the nuclear spins of carbon atoms in the proteins, NMR measures a trait known as chemical shift for some of the individual atoms in the sample, which reveals how those atoms are connected.
“You look at changes in chemical shift and that tells you, for example, if there is an alpha helix or a beta sheet, which are two different conformations that a protein backbone often takes,” Frederick says.
However, traditional NMR is not very sensitive, because it only allows you to get information from about 0.001 percent of the carbon nuclei in a sample.
For the past 20 years, Griffin’s lab has been developing dynamic nuclear polarization, which requires transferring polarization from electrons to carbon nuclei, using microwaves generated by a gyrotron. This allows researchers to gain information from 4 percent of the carbon atoms, a significant boost in sensitivity.
With traditional solid-state NMR, about 30 milligrams of purified protein are needed to get information within a reasonable amount of time. But with the boost in sensitivity from DNP, the protein no longer needs to be purified, and it can be analyzed in quantities that would normally be found inside a cell.
“It’s opening up a completely new set of problems that we can access,” Frederick says. “Using the sensitivity enhancement technique allows you to look at the protein at the correct levels, which is really important when you’re thinking about its biology.”
To make sure they are getting data only on the protein of interest, the researchers label their target protein with a radioactive form of carbon known as carbon-13, while the rest of the proteins are unlabeled.
“This technique has the potential to really open up a wide range of studies, under physiological conditions, that no one has been able to do before,” says Tim Cross, director of the NMR program at the National High Magnetic Field Laboratory and a professor at Florida State University.
“You don’t have to crystallize the proteins, you don’t have to put them into a uniform solution. You can study them in their natural environment, and that’s tremendously exciting,” says Cross, who was not part of the research team.
Protein folding
In the Cell paper, the researchers analyzed a yeast protein called Sup35, which Lindquist’s lab has been studying for many years. Sup35 is a prion — a type of protein that can form tangled clumps known as amyloids. Its usual function is to help cells terminate protein translation, but when it takes on an amyloid structure, it stops performing that function.
These are both normal states for the yeast protein, but when human proteins form amyloids they are usually associated with diseases — especially neurodegenerative diseases such as Alzheimer’s, Parkinson’s, and Huntington’s, as well as rheumatoid arthritis.
In previous studies using traditional NMR with purified Sup35 protein, scientists have found that a large section of the protein, which forms the amyloid, has a beta-sheet structure, which resembles the folds of an accordion. Another large section is intrinsically disordered and tends to flop around instead of forming a consistent structure.
However, in the new study, when the researchers looked at Sup35 surrounded by other cellular proteins, they found that the intrinsically disordered region actually does take on a regular structure, which they believe to be beta sheets. However, more powerful resolution is needed to tell for sure, Frederick says.
Frederick plans to continue using this NMR technique to study other yeast proteins, as well as human amyloid proteins. In particular, she wants to study why proteins can take on different conformations in different types of cells, or cells from people with different genetic backgrounds.
* * *
Susan Lindquist’s primary affiliation is with Whitehead Institute for Biomedical Research, where her laboratory is located and all her research is conducted. She is also a Howard Hughes Medical Institute investigator and a professor of biology at Massachusetts Institute of Technology.
* * *
Citation
Frederick, K. K., Michaelis, V. K., Corzilius, B., Ong, T. C., Jacavone, A. C., Griffin, R. G., & Lindquist, S. (2015). Sensitivity-enhanced NMR reveals alterations in protein structure by cellular milieus. Cell, 163(3), 620-628.
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu



