Rumors of Male Chromosome's Demise Greatly Exaggerated, Study Finds
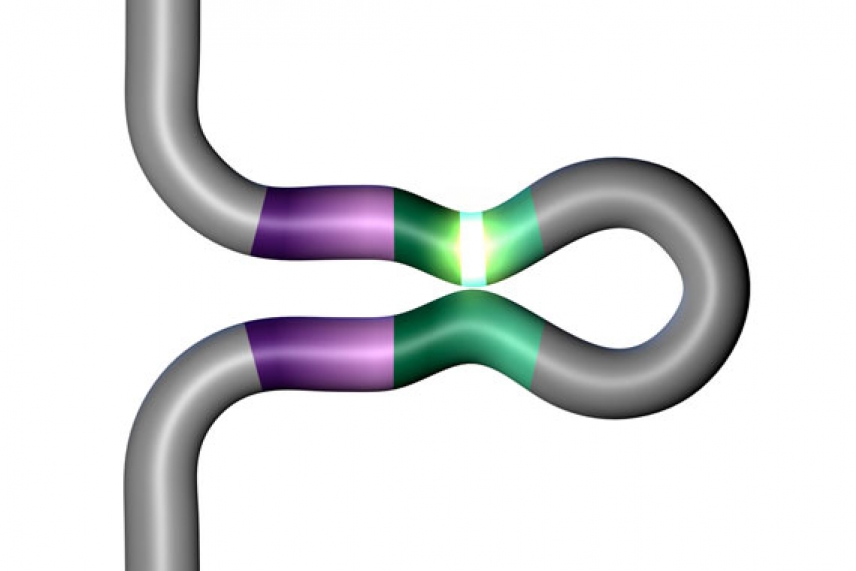
The X and Y chromosomes swap parts only at their tips.
Credit: Howard Hughes Medical Institute
CAMBRIDGE, Mass. — In the biological battle between the sexes, the Y chromosome has suffered defeat after defeat. The male-determining chromosome has seen its gene supply shrink from more than 1,000 genes when sex chromosomes first evolved, to what scientists once thought was only a handful of genes, a downward trend predicted to continue until the Y disappeared altogether.
But two studies presented today at a Washington, D.C., press conference and published in this week’s issue of the journal Nature suggest that the rumors of the Y’s demise have been greatly exaggerated. Researchers from Whitehead Institute for Biomedical Research in Cambridge, Mass., and Washington University School of Medicine in St. Louis found that not only does the Y contain far more genes than scientists thought—the team found about 78 genes—it also includes a large number of genes arranged in pairs along this single chromosome in ways that may allow the Y to mimic the paired chromosome structure of the rest of the genome.
The findings, involving observations in both human and chimp male chromosomes, could explain how the Y repairs injured genes without the benefit of sexual recombination—the method of gene repair used by all other chromosomes. It’s an elegant system that would debunk the theory of a “rotting Y”—the widely held notion that the male chromosome and its dead or dying genes will continue to rot away over the next 5 million years until there’s nothing left.
“We have a new way of understanding how the rotting tendencies of the Y are counteracted,” said David Page, a scientist at Whitehead Institute and lead researcher on this project. Page also holds an appointment as an investigator with Howard Hughes Medical Institute.
All chromosomes in the nucleus come in pairs—except the Y. Each member of a chromosomal pair draws on its mate for genetic repair through sexual recombination. When one half suffers a genetic injury, as is the case with many diseases, it can discard the mutated gene and replace it with a normal copy drawn from the other member of the pair. But the Y has no sexual “partner” with which to swap out defective genes.
“Genes constantly are being bombarded with little injuries—mutations. Mutations can either be beneficial or detrimental, but they are far more often detrimental,” said Page. “On the Y, detrimental mutations cannot be discarded.”
There’s no question that this inability to discard has cost the Y hundreds of genes over time. Many of the chromosome’s genes either have weakened or died out altogether. Sexual recombination is a card game the Y just can’t win. But this new research suggests it doesn’t always need to. For critical genes, it swaps with itself.
“This study shows that the Y chromosome has become very efficient at preserving its important genes,” said co-lead investigator Richard K. Wilson, director of the Genome Sequencing Center at Washington University School of Medicine in St. Louis. “It’s found different ways to do the things chromosomes must do to evolve, survive and thrive.”
Key to these findings is that researchers identified this unique gene repair technique not only in a human Y chromosome, but also in a chimp Y.
“When we look at the human Y, compared with the chimp Y,” Page said, “what we can infer is that during the last 5 million years, since we and chimps parted company, this overwriting of one gene copy by another has been going on frequently in our Y chromosome and in the chimp Y chromosome.”
While it’s true that over millions of years the male sex chromosome has lost hundreds of genes and seen many others crippled, the biggest concern has been gene health in the regions of the Y that control sperm production. Now, it seems that the intricate outlay of duplicate gene sequences in the area responsible for sperm production could prevent the erosion of these critically important genes.
“These papers and these findings show that while there is a slow process of degradation, there has been a concurrent gain on the Y in genes that allow for spermatogenesis,” said Robert Waterston, now a scientist at the University of Washington who was a lead researcher at the Genome Sequencing Center at Washington University during these studies. “The Y has found a way to keep these genes coherent despite a rather unstable structure. That instability of structure could be disastrous for a particular individual, but it won’t be disastrous for the Y, because the deleted Ys would not be passed on.”
For the study, researchers mapped the gene sequence of a Y chromosome from an anonymous male, as well as parts of a Y chromosome from a chimpanzee. This technically challenging process involved delicately unwrapping the two arms on each of the eight palindromes the scientists discovered, and analyzing the near-identical gene sequences inside.
“Most chromosomes are like a typical thousand-piece jigsaw puzzle—a pretty picture split into pieces with easily identifiable markings,” said Wilson. “The Y chromosome, on the other hand, was like a picture of a small sailboat on the ocean with lots of blue sky, no clouds and hundreds of pieces that looked exactly alike. Determining exactly where each piece went in the grand scheme required a lot of work.”
Studies of the Y chromosome in humans and other species haven’t always caught the collective eye of research biologists. In fact, the Y chromosome has not been studied in comparable detail in any other species.
But the small number of people interested in the Y has steadily increased in the last few years. Today, the field is populated with researchers interested in understanding problems related to infertility, the mystery surrounding the origins of modern populations, and other projects in which the Y chromosome is implicated. For scientists like Scott Hawley, a biologist with the Stowers Institute for Medical Research in Kansas City, Mo., who studies chromosomal pairing, these findings open new doors.
“I thought about, talked about, wrote about the Y as a rotting chromosome that really only had one important gene—the one that determines sex,” Hawley said. “But this makes perfectly good sense. It’s one of those ‘Ah-ha!’ experiences that, after you hear it, you think, ‘It had to be that way. Why didn’t we think of this before?’ It’s just revolutionary work.”
The next steps in this research will involve a closer scrutiny of this self-repair mechanism, as well as plans to sequence the Y chromosome in mice.
“This analysis of the Y chromosome shows how the data produced by the Human Genome Project will continue to generate unexpected but fascinating results for years to come,” said Francis S. Collins, director of the National Human Genome Research Institute. “Unless the human sequence had been finished to the highest level of accuracy, the Y chromosome researchers would not have been able to identify the Y’s unusual genetic structure and the novel mechanism for maintaining its integrity.”
This research was supported with funds from the National Human Genome Research Institute and the Howard Hughes Medical Institute.
***
Skaletsky, H., et al. (2003). “The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes.” Nature 423 (6942), 825-837.
Rosen, S., et al. (2003). “Abundant gene conversion between arms of palindromes in human and ape Y chromosomes.” Nature 423 (6942), 873-876.
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu


