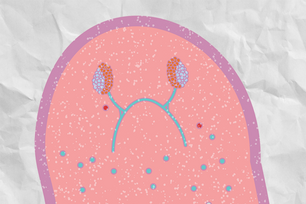
Planarians offer a better view of eye development

Planarian with eye spots (top) and one missing eye spots (bottom) due to repression of the ovo gene
Courtesy of Cell Reports, S. Lapan, P. Reddien.
CAMBRIDGE, Mass. – Planarian flatworms have come under intense study for their renowned ability to regenerate any missing body part, even as adults. But now they may take on a starring role as a model system for studying eye development and eye diseases in vertebrates, including humans.
This expansion of the planarian job description comes courtesy of Whitehead Institute researchers, who this week are publishing in Cell Reports an exhaustive catalog of genes active in the planarian eye.
“It’s exciting to get this complete list of genes in one fell swoop,” says Whitehead Member Peter Reddien, who is also a Howard Hughes Medical Institute (HHMI) Early Career Scientist and an associate professor of biology at Massachusetts Institute of Technology. “This provides perhaps the most comprehensive list of genes involved in eye biology in a model system other than Drosophila that can be used for rapidly studying the function of those genes.”
The compound eyes of fruit flies were the most thoroughly studied invertebrate eyes, owing to the short fly life cycle and a library of well-described eye-related genes in fruit flies. Planarians present a new genetic system, with an eye based on one optic cup lined with pigment cells.
To create a list of genes showing enriched activity in the planarian eye, Sylvain Lapan, a graduate student in the Reddien lab, analyzed more than 2,000 planarian eyes. Lapan and Reddien found 600 active genes and studied 200 of them in more detail. Several of the identified genes are known to have versions that play a role in the vertebrate eye but have not been found in the fruit fly eye. Among these are genes involved in eye development and others associated with age-related macular degeneration and Usher syndrome, a disorder that causes progressive retinal degradation.
One of the key genes identified in planarian eye development is the transcription factor ovo, which activates the expression of many other genes as the eye forms. Until now ovo had been associated with neural tube and germ cell development in other organisms, but not with the eye. In planarians, ovo is vital for eye for regeneration and eye maintenance in the adult, and is also active in eye development in the embryo. In fact, when ovo is experimentally turned off, planarians with head amputations cannot regenerate their eyes and eyes of otherwise normal adult planarians vanish after a couple months.
“Similar mechanisms are used to make eyes during homeostasis, regeneration, and embryonic development in planarians,” says Lapan. “We now know way more about genes that regulate eye formation in these animals than for any invertebrate other than Drosophila. Planarian eyes are very different from fly eyes, and we're already seeing the benefits of studying diverse model species, like the discovery of a critical role for ovo.”
This work was supported by the National Institutes of Health (NIH) and the Keck Foundation.
* * *
Peter Reddien’s primary affiliation is with Whitehead Institute for Biomedical Research, where his laboratory is located and all his research is conducted. He is also a Howard Hughes Medical Institute Early Career Scientist and an associate professor of biology at Massachusetts Institute of Technology.
* * *
Citation:
Lapan, S., & Reddien, P. (2012). Transcriptome Analysis of the Planarian Eye Identifies ovo as a Specific Regulator of Eye Regeneration. Cell Reports, 2(2), 294-307. doi:10.1016/j.celrep.2012.06.018
Contact
Communications and Public Affairs
Phone: 617-452-4630
Email: newsroom@wi.mit.edu